bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2156_orf1
Length=368
Score E
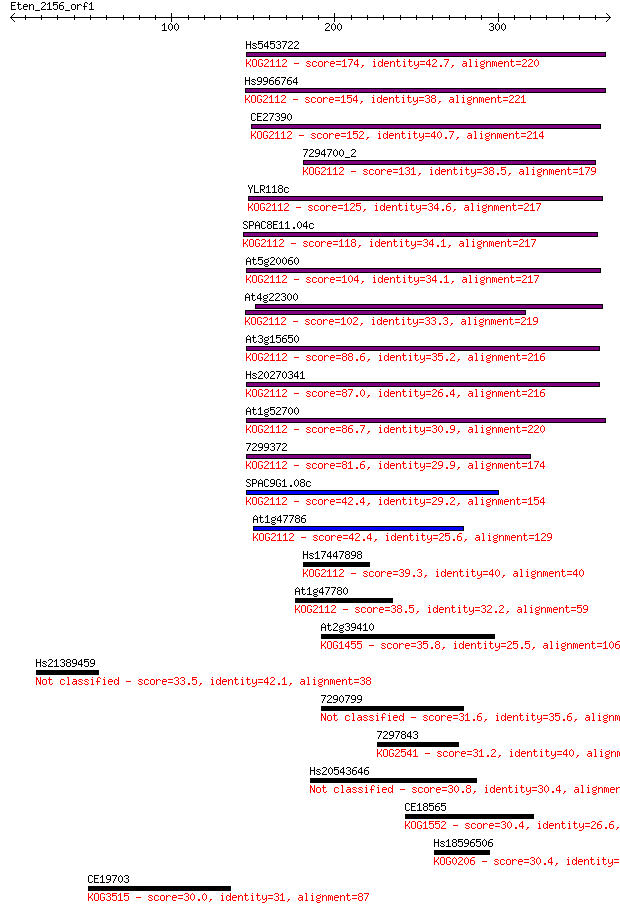
Sequences producing significant alignments: (Bits) Value
Hs5453722 174 3e-43
Hs9966764 154 3e-37
CE27390 152 1e-36
7294700_2 131 2e-30
YLR118c 125 2e-28
SPAC8E11.04c 118 2e-26
At5g20060 104 3e-22
At4g22300 102 1e-21
At3g15650 88.6 2e-17
Hs20270341 87.0 6e-17
At1g52700 86.7 7e-17
7299372 81.6 2e-15
SPAC9G1.08c 42.4 0.001
At1g47786 42.4 0.002
Hs17447898 39.3 0.014
At1g47780 38.5 0.025
At2g39410 35.8 0.13
Hs21389459 33.5 0.80
7290799 31.6 2.9
7297843 31.2 4.2
Hs20543646 30.8 5.1
CE18565 30.4 5.7
Hs18596506 30.4 6.2
CE19703 30.0 8.7
> Hs5453722
Length=230
Score = 174 bits (441), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 94/221 (42%), Positives = 129/221 (58%), Gaps = 14/221 (6%)
Query 146 KATGTLIFMHGLGDTAGGWTELLQLFEAAQIHEKARLILPTAPTRPVTLNMGVRMTAWSD 205
KAT +IF+HGLGDT GW E ++ I + I P AP RPVTLNM V M +W D
Sbjct 19 KATAAVIFLHGLGDTGHGWAEAFAGIRSSHI----KYICPHAPVRPVTLNMNVAMPSWFD 74
Query 206 IRGLSADSSEDKDGLMASKARIDRIIDSEIKAGIHPSRILVGGFSQGGALAYLVGLTSPH 265
I GLS DS ED+ G+ + I +ID E+K GI +RI++GGFSQGGAL+ LT+
Sbjct 75 IIGLSPDSQEDESGIKQAAENIKALIDQEVKNGIPSNRIILGGFSQGGALSLYTALTTQQ 134
Query 266 KLGGILSLSSWCPLGKEIQVSP-HYAQAVPRILHCHGAVDDIVRPIYGQTSVESVRNRLI 324
KL G+ +LS W PL P A IL CHG D +V ++G +VE ++ L+
Sbjct 135 KLAGVTALSCWLPLRASFPQGPIGGANRDISILQCHGDCDPLVPLMFGSLTVEKLKT-LV 193
Query 325 DSGARPEKCQDDITFKLYEGLGHSANQQELNDIKVFLSRIF 365
+ ++TFK YEG+ HS+ QQE+ D+K F+ ++
Sbjct 194 NPA--------NVTFKTYEGMMHSSCQQEMMDVKQFIDKLL 226
> Hs9966764
Length=231
Score = 154 bits (389), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 84/221 (38%), Positives = 122/221 (55%), Gaps = 13/221 (5%)
Query 145 SKATGTLIFMHGLGDTAGGWTELLQLFEAAQIHEKARLILPTAPTRPVTLNMGVRMTAWS 204
+ T +IF+HGLGDT W + L + + I P AP PVTLNM + M +W
Sbjct 21 ERETAAVIFLHGLGDTGHSWADALSTIRLPHV----KYICPHAPRIPVTLNMKMVMPSWF 76
Query 205 DIRGLSADSSEDKDGLMASKARIDRIIDSEIKAGIHPSRILVGGFSQGGALAYLVGLTSP 264
D+ GLS D+ ED+ G+ + I +I+ E+K GI +RI++GGFSQGGAL+ LT P
Sbjct 77 DLMGLSPDAPEDEAGIKKAAENIKALIEHEMKNGIPANRIVLGGFSQGGALSLYTALTCP 136
Query 265 HKLGGILSLSSWCPLGKEIQVSPHYAQAVPRILHCHGAVDDIVRPIYGQTSVESVRNRLI 324
H L GI++LS W PL + + + + IL CHG +D +V +G + E +R
Sbjct 137 HPLAGIVALSCWLPLHRAFPQAANGSAKDLAILQCHGELDPMVPVRFGALTAEKLR---- 192
Query 325 DSGARPEKCQDDITFKLYEGLGHSANQQELNDIKVFLSRIF 365
S P + Q FK Y G+ HS+ QE+ +K FL ++
Sbjct 193 -SVVTPARVQ----FKTYPGVMHSSCPQEMAAVKEFLEKLL 228
> CE27390
Length=223
Score = 152 bits (384), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 87/214 (40%), Positives = 119/214 (55%), Gaps = 15/214 (7%)
Query 149 GTLIFMHGLGDTAGGWTELLQLFEAAQIHEKARLILPTAPTRPVTLNMGVRMTAWSDIRG 208
GTLIF+HGLGD GW + F+ H+ + I P + RPVTLNMG+RM AW D+ G
Sbjct 20 GTLIFLHGLGDQGHGWADA---FKTEAKHDNIKFICPHSSERPVTLNMGMRMPAWFDLFG 76
Query 209 LSADSSEDKDGLMASKARIDRIIDSEIKAGIHPSRILVGGFSQGGALAYLVGLTSPHKLG 268
L ++ ED+ G+ + + ++ID+E+ AGI SRI VGGFS GGALA GLT P KLG
Sbjct 77 LDPNAQEDEQGINRATQYVHQLIDAEVAAGIPASRIAVGGFSMGGALAIYAGLTYPQKLG 136
Query 269 GILSLSSWCPLGKEIQVSPHYAQAVPRILHCHGAVDDIVRPIYGQTSVESVRNRLIDSGA 328
GI+ LSS+ + S A P L HG D +V +GQ S + ++
Sbjct 137 GIVGLSSFFLQRTKFPGSFTANNATPIFL-GHGTDDFLVPLQFGQMSEQYIK-------- 187
Query 329 RPEKCQDDITFKLYEGLGHSANQQELNDIKVFLS 362
K + Y G+ HS+ +E+ D+K FLS
Sbjct 188 ---KFNPKVELHTYRGMQHSSCGEEMRDVKTFLS 218
> 7294700_2
Length=211
Score = 131 bits (329), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 69/179 (38%), Positives = 105/179 (58%), Gaps = 12/179 (6%)
Query 181 RLILPTAPTRPVTLNMGVRMTAWSDIRGLSADSSEDKDGLMASKARIDRIIDSEIKAGIH 240
++I PTAPT+PV+LN G RM +W D++ L ED+ G+ +++ + +I EI AGI
Sbjct 17 KVICPTAPTQPVSLNAGFRMPSWFDLKTLDIGGPEDEPGIQSARDSVHGMIQKEISAGIP 76
Query 241 PSRILVGGFSQGGALAYLVGLTSPHKLGGILSLSSWCPLGKEIQVSPHYAQAVPRILHCH 300
+RI++GGFSQGGALA LT L G+++LS W PL K+ + + VP I H
Sbjct 77 ANRIVLGGFSQGGALALYSALTYDQPLAGVVALSCWLPLHKQFPGAKVNSDDVP-IFQAH 135
Query 301 GAVDDIVRPIYGQTSVESVRNRLIDSGARPEKCQDDITFKLYEGLGHSANQQELNDIKV 359
G D +V +GQ S +++ + ++TFK Y GL HS++ E++D+KV
Sbjct 136 GDYDPVVPYKFGQLSASLLKSFM-----------KNVTFKTYSGLSHSSSDDEMDDVKV 183
> YLR118c
Length=227
Score = 125 bits (313), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 75/221 (33%), Positives = 110/221 (49%), Gaps = 14/221 (6%)
Query 147 ATGTLIFMHGLGDTAGGWTELLQLFEAAQ--IHEKARLILPTAPTRPVTLNMGVRMTAWS 204
A T+IF+HGLGDT GW L Q + + + P AP VT N G M AW
Sbjct 13 ARQTIIFLHGLGDTGSGWGFLAQYLQQRDPAAFQHTNFVFPNAPELHVTANGGALMPAWF 72
Query 205 DIRGLSADSSE-DKDGLMASKARIDRIIDSEIKAGIHPSRILVGGFSQGGALAYLVGLTS 263
DI S+ D DG M S I++ + EI GI P +I++GGFSQG ALA +T
Sbjct 73 DILEWDPSFSKVDSDGFMNSLNSIEKTVKQEIDKGIKPEQIIIGGFSQGAALALATSVTL 132
Query 264 PHKLGGILSLSSWCPLGKEIQVSPHYAQAVPRILHCHGAVDDIVRPIYGQTSVESVRNRL 323
P K+GGI++LS +C + ++ + I H HG +D +V G + + +
Sbjct 133 PWKIGGIVALSGFCSIPGILKQHKNGINVKTPIFHGHGDMDPVVPIGLGIKAKQFYQ--- 189
Query 324 IDSGARPEKCQ-DDITFKLYEGLGHSANQQELNDIKVFLSR 363
+ C+ + FK+Y+G+ HS EL D+ F+ +
Sbjct 190 -------DSCEIQNYEFKVYKGMAHSTVPDELEDLASFIKK 223
> SPAC8E11.04c
Length=224
Score = 118 bits (295), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 74/219 (33%), Positives = 115/219 (52%), Gaps = 16/219 (7%)
Query 144 PSKA-TGTLIFMHGLGDTAGGWTELLQLFEAAQIHEKARLILPTAPTRPVTLNMGVRMTA 202
PS A T T+IF+HGLGD+ GW+ + + + + I P AP+ PVT+N G++M A
Sbjct 12 PSVAHTATVIFLHGLGDSGQGWSFMANTWSN---FKHIKWIFPNAPSIPVTVNNGMKMPA 68
Query 203 WSDIRGLSADSSEDKDGLMASKARIDRIIDSEIKAGIHPSRILVGGFSQGGALAYLVGLT 262
W DI + ED++G++ S ++ +ID+E+ GI RIL+GGFSQG ++ GLT
Sbjct 69 WYDIYSFADMKREDENGILRSAGQLHELIDAELALGIPSDRILIGGFSQGCMVSLYAGLT 128
Query 263 SPHKLGGILSLSSWCPLGKEIQVS-PHYAQAVPRILHCHGAVDDIVRPIYGQTSVESVRN 321
P +L GI+ S + PL + + A+ +P IL + D IV + S + + N
Sbjct 129 YPKRLAGIMGHSGFLPLASKFPSALSRVAKEIP-ILLTYMTEDPIVPSVLSSASAKYLIN 187
Query 322 RLIDSGARPEKCQDDITFKLYEGLGHSANQQELNDIKVF 360
L KC D + +EG HS + + + F
Sbjct 188 NL------QLKCLD----RPFEGDAHSLSSESFMAMYKF 216
> At5g20060
Length=252
Score = 104 bits (260), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 74/237 (31%), Positives = 117/237 (49%), Gaps = 42/237 (17%)
Query 146 KATGTLIFMHGLGDTAGGWTELLQLFEAAQIHEKARLILPTAPTRPVTLNMGVRMTAWSD 205
K T++++HGLGD W++LL+ I + I PTAP++P++L G TAW D
Sbjct 31 KHQATIVWLHGLGDNGSSWSQLLETLPLPNI----KWICPTAPSQPISLFGGFPSTAWFD 86
Query 206 IRGLSADSSEDKDGLMASKARIDRIIDSE---IKAGIHPSRILVGGFSQGGALA------ 256
+ ++ D +D +GL + A + ++ +E IK G VGGFS G A +
Sbjct 87 VVDINEDGPDDMEGLDVAAAHVANLLSNEPADIKLG-------VGGFSMGAATSLYSATC 139
Query 257 -----YLVGLTSPHKLGGILSLSSWCPLGK------EIQVSPHYAQAVPRILHCHGAVDD 305
Y G P L I+ LS W P K E + + A ++P I+ CHG DD
Sbjct 140 FALGKYGNGNPYPINLSAIIGLSGWLPCAKTLAGKLEEEQIKNRAASLP-IVVCHGKADD 198
Query 306 IVRPIYGQTSVESVRNRLIDSGARPEKCQDDITFKLYEGLGHSANQQELNDIKVFLS 362
+V +G+ S ++ L+ +G + +TFK Y LGH QEL+++ +L+
Sbjct 199 VVPFKFGEKSSQA----LLSNGFK------KVTFKPYSALGHHTIPQELDELCAWLT 245
> At4g22300
Length=471
Score = 102 bits (254), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 71/224 (31%), Positives = 107/224 (47%), Gaps = 23/224 (10%)
Query 151 LIFMHGLGDTAGGWTELLQLFEAAQIHEKARLILPTAPTRPVTLNMGVRMTAWSDIRGL- 209
++++HGLGD+ + F+++++ A + P+AP PVT N G M +W D+ L
Sbjct 6 ILWLHGLGDSGPANEPIQTQFKSSEL-SNASWLFPSAPFNPVTCNNGAVMRSWFDVPELP 64
Query 210 -SADSSEDKDGLMASKARIDRIIDSEIKAGIHPSRILVGGFSQGGALAYLVGLTSPHKLG 268
S D+ ++ + + IID EI G +P + + G SQGGAL L P LG
Sbjct 65 FKVGSPIDESSVLEAVKNVHAIIDQEIAEGTNPENVFICGLSQGGALTLASVLLYPKTLG 124
Query 269 GILSLSSWCPLGKE-IQVSPHYAQAVPR--------ILHCHGAVDDIVRPIYGQTSVESV 319
G LS W P I P A+ VP IL HG D +V GQ ++ +
Sbjct 125 GGAVLSGWVPFTSSIISQFPEEAKKVPHLCFLINTPILWSHGTDDRMVLFEAGQAALPFL 184
Query 320 RNRLIDSGARPEKCQDDITFKLYEGLGHSANQQELNDIKVFLSR 363
+ ++G E FK Y GLGHS + +EL I+ ++ R
Sbjct 185 K----EAGVTCE-------FKAYPGLGHSISNKELKYIESWIKR 217
Score = 92.4 bits (228), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 58/174 (33%), Positives = 82/174 (47%), Gaps = 3/174 (1%)
Query 145 SKATGTLIFMHGLGDTAGGWTELLQLFEAAQIHEKARLILPTAPTRPVTLNMGVRMTAWS 204
S A ++++HGLGD+ + LF + Q + + P+AP PV+ N G M +W
Sbjct 285 SMARTFILWLHGLGDSGPANEPIKTLFRS-QEFRNTKWLFPSAPPNPVSCNYGAVMPSWF 343
Query 205 DIRGL--SADSSEDKDGLMASKARIDRIIDSEIKAGIHPSRILVGGFSQGGALAYLVGLT 262
DI L +A S +D+ L+ + + IID EI GI+P + + GFSQGGAL L
Sbjct 344 DIPELPLTAGSPKDESSLLKAVKNVHAIIDKEIAGGINPENVYICGFSQGGALTLASVLL 403
Query 263 SPHKLGGILSLSSWCPLGKEIQVSPHYAQAVPRILHCHGAVDDIVRPIYGQTSV 316
P +GG S W P I IL HG D V GQ ++
Sbjct 404 YPKTIGGGAVFSGWIPFNSSITNQFTEDAKKTPILWSHGIDDKTVLFEAGQAAL 457
> At3g15650
Length=255
Score = 88.6 bits (218), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 76/236 (32%), Positives = 110/236 (46%), Gaps = 41/236 (17%)
Query 146 KATGTLIFMHGLGDTAGGWTELLQLFEAAQIHEKARLILPTAPTRPVTLNMGVRMTAWSD 205
K T++++HGLGD ++LL+ I + I PTAP+RPV+L G TAW D
Sbjct 31 KHQATIVWLHGLGDNGSSSSQLLESLPLPNI----KWICPTAPSRPVSLLGGFPCTAWFD 86
Query 206 IRGLSADSSEDKDGLMASKARIDRIIDSE---IKAGIHPSRILVGGFSQGGALA------ 256
+ +S D +D +GL AS A I ++ +E +K GI GGFS G A+A
Sbjct 87 VGEISEDLHDDIEGLDASAAHIANLLSAEPTDVKVGI-------GGFSMGAAIALYSTTC 139
Query 257 -----YLVGLTSPHKLGGILSLSSWCPLGKEIQVSPHYAQAVPR------ILHCHGAVDD 305
Y G L + LS W P + ++ + V R IL HG DD
Sbjct 140 YALGRYGTGHAYTINLRATVGLSGWLPGWRSLRSKIESSNEVARRAASIPILLAHGTSDD 199
Query 306 IVRPIYGQTSVESVRNRLIDSGARPEKCQDDITFKLYEGLGHSANQQELNDIKVFL 361
+V +G+ S S L +G R FK YEGLGH +E++++ +L
Sbjct 200 VVPYRFGEKSAHS----LAMAGFR------QTMFKPYEGLGHYTVPKEMDEVVHWL 245
> Hs20270341
Length=237
Score = 87.0 bits (214), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 57/220 (25%), Positives = 104/220 (47%), Gaps = 17/220 (7%)
Query 146 KATGTLIFMHGLGDTAGG---WTELLQLFEAAQIHEKARLILPTAPTRPVTLNMGVRMTA 202
+ + +LIF+HG GD+ G W + Q+ + ++I PTAP R T G
Sbjct 19 RHSASLIFLHGSGDSGQGLRMWIK--QVLNQDLTFQHIKIIYPTAPPRSYTPMKGGISNV 76
Query 203 WSDIRGLSADSSEDKDGLMASKARIDRIIDSEIKAGIHPSRILVGGFSQGGALAYLVGLT 262
W D ++ D E + + + +ID E+K+GI +RIL+GGFS GG +A +
Sbjct 77 WFDRFKITNDCPEHLESIDVMCQVLTDLIDEEVKSGIKKNRILIGGFSMGGCMAMHLAYR 136
Query 263 SPHKLGGILSLSSWCPLGKEIQVSPHYAQAV-PRILHCHGAVDDIVRPIYGQTSVESVRN 321
+ + G+ +LSS+ + + + V P + CHG D++V + + + N
Sbjct 137 NHQDVAGVFALSSFLNKASAVYQALQKSNGVLPELFQCHGTADELVLHSWAEET-----N 191
Query 322 RLIDSGARPEKCQDDITFKLYEGLGHSANQQELNDIKVFL 361
++ S K F + + H ++ EL+ +K+++
Sbjct 192 SMLKSLGVTTK------FHSFPNVYHELSKTELDILKLWI 225
> At1g52700
Length=223
Score = 86.7 bits (213), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 68/231 (29%), Positives = 103/231 (44%), Gaps = 55/231 (23%)
Query 146 KATGTLIFMHGLGDTAGGWTELLQLFEAAQIHEKARLILPTAPTRPVTLNMGVRMTAWSD 205
K TL+++HGLGD ++L+ I + I PTAP+RPVT G TAW D
Sbjct 31 KHQATLVWLHGLGDNGSSSSQLMDSLHLPNI----KWICPTAPSRPVTSLGGFTCTAWFD 86
Query 206 IRGLSADSSEDKDGLMASKARIDRIIDSEIKAGIHPSRILVGGFSQGGALA--------- 256
+ +S D +D +GL AS + I ++ SE P+ + +GGFS G A++
Sbjct 87 VGEISEDGHDDLEGLDASASHIANLLSSE------PADVGIGGFSMGAAISLYSATCYAL 140
Query 257 --YLVGLTSPHKLGGILSLSSWCPLGKEIQVSPHYAQAVPRILHCHGAVDDIVRPIYGQT 314
Y G P L ++ LS W P DD+V +G+
Sbjct 141 GRYGTGHAYPINLQAVVGLSGWLP------------------------ADDVVPYRFGEK 176
Query 315 SVESVRNRLIDSGARPEKCQDDITFKLYEGLGHSANQQELNDIKVFLSRIF 365
S +S L +G R FK YEGLGH +E++++ +L+ +
Sbjct 177 SAQS----LGMAGFRLA------MFKPYEGLGHYTVPREMDEVVHWLTTML 217
> 7299372
Length=235
Score = 81.6 bits (200), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 52/177 (29%), Positives = 88/177 (49%), Gaps = 3/177 (1%)
Query 146 KATGTLIFMHGLGDTAGGWTELLQLFEAAQI-HEKARLILPTAPTRPVTLNMGVRMTAWS 204
K T ++IF HG GDT E ++ + + ++I PTAP + T G W
Sbjct 13 KHTASVIFFHGSGDTGPNVLEWVRFLIGRNLEYPHIKIIYPTAPKQKYTPLDGELSNVWF 72
Query 205 DIRGLSADSSEDKDGLMASKARIDRIIDSEIKAGIHPSRILVGGFSQGGALAYLVGLTSP 264
D + ++ +SE K + ++++ID E+ +GI +RI+VGGFS GGALA G
Sbjct 73 DRKSVNIAASESKKSMSQCYDAVNQLIDEEVASGIPLNRIVVGGFSMGGALALHTGYHLR 132
Query 265 HKLGGILSLSSWCPLGKEI--QVSPHYAQAVPRILHCHGAVDDIVRPIYGQTSVESV 319
L G+ + SS+ G + ++ ++ P + HG D +V +G + E++
Sbjct 133 RSLAGVFAHSSFLNRGSVVYDSLANGKDESFPELRMYHGERDTLVPKDWGLETFENL 189
> SPAC9G1.08c
Length=241
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 45/168 (26%), Positives = 67/168 (39%), Gaps = 26/168 (15%)
Query 146 KATGTLIFMHGLGDTAGGWTELLQ---LFEAAQIHEKARLILPTAPTRPVTLNMGVRMTA 202
K +I MHGLGD+ + + + L + I + LP P M
Sbjct 22 KVHNVVILMHGLGDSHKSFANMAKNVPLPNTSYISLRGPYRLPLDFENPGGNWM------ 75
Query 203 WSDIRGLSADSSEDKDGLMASKAR-------IDRIIDSEIKAGIHPSRILVGGFSQGG-- 253
W + D D++G + S+A I +I + + GI SRI GF QG
Sbjct 76 WGE------DVHFDQNGELQSEADFSKSFTMISNLIGNLLSYGILSSRIFFFGFGQGAMV 129
Query 254 ALAYLVGLTSPHKLGGILSLSSWCPLGKEIQVSPHYAQA--VPRILHC 299
AL L++ ++LGGI S PL + P + + LHC
Sbjct 130 ALYSCYKLSTKYQLGGIFSFGGTLPLSITLPNHPFHVPVYLFEKRLHC 177
> At1g47786
Length=186
Score = 42.4 bits (98), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 33/137 (24%), Positives = 54/137 (39%), Gaps = 19/137 (13%)
Query 150 TLIFMHGLGDTAGGWTELLQLFEAAQIHEKARLILPTAPTRPVTLNMGVRMTAWSDIRGL 209
T++++H +G+T+ + I + I PTAP RPVT+ G+ AW DI +
Sbjct 58 TIVWLHDIGETSANSVRFARQLGLRNI----KWICPTAPRRPVTILGGMETNAWFDIAEI 113
Query 210 SADSSEDKDGLMASKARIDRIIDSEIKAGIHPSRILVGGFSQGGALAYLVGLTSPHKLGG 269
S + +D+ L + I + I GG G A A + S +
Sbjct 114 SENMQDDEVSLHHAALSIANLFSDHASPNI-------GGMGMGAAQALYLASKSCYDTNQ 166
Query 270 --------ILSLSSWCP 278
++ L W P
Sbjct 167 RLQIKPRVVIGLKGWLP 183
> Hs17447898
Length=141
Score = 39.3 bits (90), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 181 RLILPTAPTRPVTLNMGVRMTAWSDIRGLSADSSEDKDGL 220
+ I P P+TLNM + +++W D+ GLS DS E++ G+
Sbjct 101 KCICPHVLIMPMTLNMNMAISSWFDVFGLSPDSQEEEPGI 140
> At1g47780
Length=126
Score = 38.5 bits (88), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 31/59 (52%), Gaps = 0/59 (0%)
Query 176 IHEKARLILPTAPTRPVTLNMGVRMTAWSDIRGLSADSSEDKDGLMASKARIDRIIDSE 234
+++ + I PTAP RP+T+ G+ AW DI LS + +D L + I ++ E
Sbjct 20 MNKNVKWICPTAPRRPLTILGGMETNAWFDIAELSENMQDDVASLNHAALSIANLLSEE 78
> At2g39410
Length=311
Score = 35.8 bits (81), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 27/117 (23%), Positives = 53/117 (45%), Gaps = 12/117 (10%)
Query 192 VTLNMGVR--MTAWSDIRGLSADSSEDKDGLMASKARIDRIIDS---------EIKAGIH 240
+T+N R + A + G+ + DGL A + DR++D E +
Sbjct 45 ITMNSTARRLVKAGFAVYGMDYEGHGKSDGLSAYISNFDRLVDDVSTHYTAICEREENKW 104
Query 241 PSRILVGGFSQGGALAYLVGLTSPHKLGGILSLSSWCPLGKEIQVSPHYAQAVPRIL 297
R ++G S GGA+ L+G +P G + ++ C + +E++ SP + +++
Sbjct 105 KMRFMLGE-SMGGAVVLLLGRKNPDFWDGAILVAPMCKIAEEMKPSPFVISILTKLI 160
> Hs21389459
Length=281
Score = 33.5 bits (75), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 17 QLVGILVLLAAQSTVRSSAFRAFGPLACEKRIFRQNLT 54
QL+G +LLA R SAFR+ GPL + ++ LT
Sbjct 189 QLLGDELLLAKLPPSRESAFRSLGPLEAQDSLYNSPLT 226
> 7290799
Length=185
Score = 31.6 bits (70), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 31/108 (28%), Positives = 45/108 (41%), Gaps = 21/108 (19%)
Query 192 VTLNMGVRMTAWSD--IRGLSADSSEDKDGLMASKARIDRIIDSEI--------KAGIHP 241
+ L RM +SD + LS+ S ++ D M K + +SE+ KA
Sbjct 37 LELQSNKRMLYFSDGVMEELSSGSEDEADAEMGDKCYDVHLNESEMPLGPRLRYKASRMG 96
Query 242 SRILVGGFSQGGALAYLVGLTSPH-----------KLGGILSLSSWCP 278
+R L G GG LA+L+G+TS K G L +W P
Sbjct 97 NRFLAGIDYVGGGLAHLLGITSSKYASELENYHRAKEHGDEDLDNWHP 144
> 7297843
Length=288
Score = 31.2 bits (69), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 31/52 (59%), Gaps = 2/52 (3%)
Query 226 RIDRIIDSEIKAG-IHPSRILVGGFSQGGALAYLVGLTSP-HKLGGILSLSS 275
++D++ D + G +HP I+V G+SQGG LA + P H + +SLSS
Sbjct 73 QVDQVRDYLNEVGKLHPEGIIVLGYSQGGLLARAAIQSLPEHNVKTFISLSS 124
> Hs20543646
Length=768
Score = 30.8 bits (68), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 31/106 (29%), Positives = 48/106 (45%), Gaps = 4/106 (3%)
Query 185 PTAPTRPVTLNMGVRMTAWSDIRGLSADSSEDKDGLMASKARI-DRIIDSEIKAGIHPS- 242
P AP + + TA S I G+S S+ K G + R +R D+ +K G HP
Sbjct 284 PAAPGQLYSFCPQALRTALSPIEGMSGRGSQPKPGSKEAPLRTQNRGEDAAVKLGGHPEA 343
Query 243 -RILVG-GFSQGGALAYLVGLTSPHKLGGILSLSSWCPLGKEIQVS 286
R+ G S+G + LV L P G +++ LG ++ +S
Sbjct 344 PRLRFGREASEGLGVGTLVALGGPPPPKGKTAVAQDLLLGADLSLS 389
> CE18565
Length=305
Score = 30.4 bits (67), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 21/94 (22%), Positives = 40/94 (42%), Gaps = 15/94 (15%)
Query 243 RILVGGFSQGGALAYLVGLTSPHKLGGILSLSSWCPLGKEIQVSPHYAQAV--------- 293
+I+V G+S G A + T+P +L G++ ++ + + P
Sbjct 151 KIVVMGYSIGTTAAVDLAATNPDRLAGVVLIAPFTSGLRLFSSKPDKPDTCWADSFKSFD 210
Query 294 ------PRILHCHGAVDDIVRPIYGQTSVESVRN 321
R+L CHG VD+++ +G E ++N
Sbjct 211 KINNIDTRVLICHGDVDEVIPLSHGLALYEKLKN 244
> Hs18596506
Length=950
Score = 30.4 bits (67), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 22/36 (61%), Gaps = 2/36 (5%)
Query 261 LTSPHKLGGILSLSSWCPLGKEIQVSP--HYAQAVP 294
L PH+LG +L SSW PL +++ + P +Y +P
Sbjct 898 LICPHQLGLLLPSSSWLPLPEDLGLGPPRYYHYFIP 933
> CE19703
Length=1270
Score = 30.0 bits (66), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 27/92 (29%), Positives = 41/92 (44%), Gaps = 5/92 (5%)
Query 49 FRQNLTA---PTAVRWPVRAE-TPRQRAPLETSARRSVLV-RLLSCTCLTRYPLRQTSSV 103
F++NLT P W R P AR +VL + L T Y L T+SV
Sbjct 708 FKENLTVRGNPAISLWQWRKNGVPFDHTIGRVFARGAVLSGKQLLSTDAGVYTLTATNSV 767
Query 104 GSAGLGLQGSSSVKGVRTAVKMPVVLSPGDTI 135
GS + ++ + T++ PV+ + GDT+
Sbjct 768 GSTNITIKLAVEYSARITSISTPVIAATGDTV 799
Lambda K H
0.320 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9050455788
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40