bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2162_orf1
Length=146
Score E
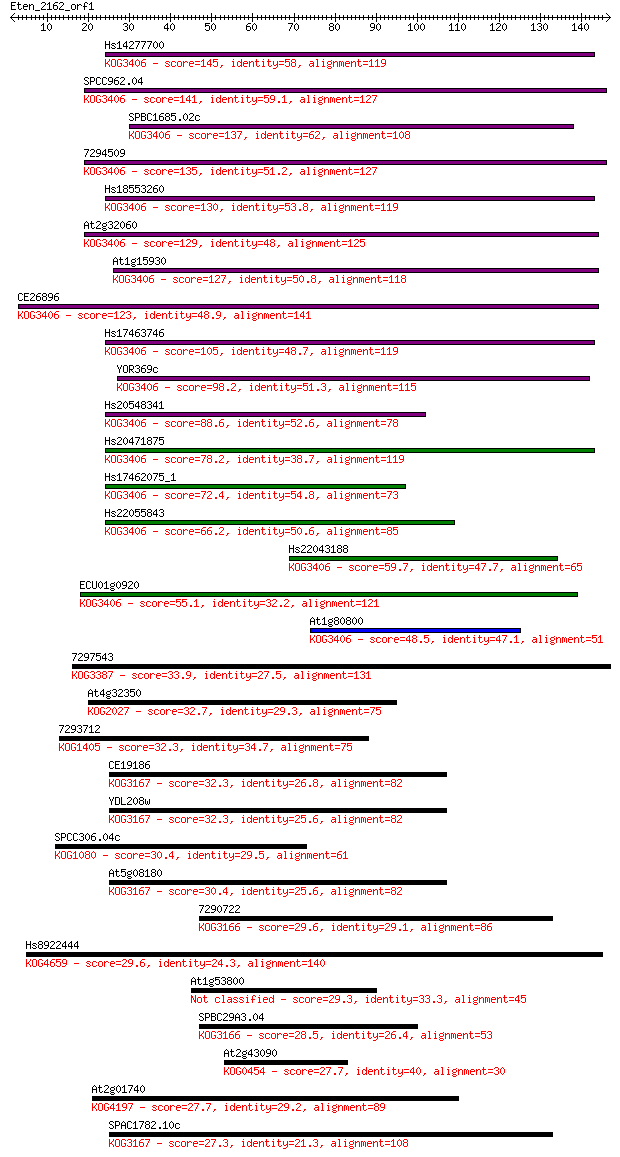
Sequences producing significant alignments: (Bits) Value
Hs14277700 145 3e-35
SPCC962.04 141 5e-34
SPBC1685.02c 137 1e-32
7294509 135 2e-32
Hs18553260 130 1e-30
At2g32060 129 2e-30
At1g15930 127 9e-30
CE26896 123 1e-28
Hs17463746 105 2e-23
YOR369c 98.2 5e-21
Hs20548341 88.6 4e-18
Hs20471875 78.2 6e-15
Hs17462075_1 72.4 2e-13
Hs22055843 66.2 2e-11
Hs22043188 59.7 2e-09
ECU01g0920 55.1 5e-08
At1g80800 48.5 4e-06
7297543 33.9 0.10
At4g32350 32.7 0.26
7293712 32.3 0.29
CE19186 32.3 0.32
YDL208w 32.3 0.37
SPCC306.04c 30.4 1.2
At5g08180 30.4 1.3
7290722 29.6 2.0
Hs8922444 29.6 2.2
At1g53800 29.3 2.5
SPBC29A3.04 28.5 4.9
At2g43090 27.7 8.0
At2g01740 27.7 8.6
SPAC1782.10c 27.3 10.0
> Hs14277700
Length=132
Score = 145 bits (365), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 69/119 (57%), Positives = 88/119 (73%), Gaps = 0/119 (0%)
Query 24 VKDLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEPAYKKLVEGLCK 83
V D+ TA++ VLK ALIHDGLARG+ E AKALD ++A C LA +C EP Y KLVE LC
Sbjct 11 VMDVNTALQEVLKTALIHDGLARGIREAAKALDKRQAHLCVLASNCDEPMYVKLVEALCA 70
Query 84 EHGIPLIEVADSKELGQWAGLCRIDNEGAARKVVGASCVAVTDFGEQSEALIFLQNHIK 142
EH I LI+V D+K+LG+W GLC+ID EG RKVVG SCV V D+G++S+A ++ + K
Sbjct 71 EHQINLIKVDDNKKLGEWVGLCKIDREGKPRKVVGCSCVVVKDYGKESQAKDVIEEYFK 129
> SPCC962.04
Length=145
Score = 141 bits (355), Expect = 5e-34, Method: Compositional matrix adjust.
Identities = 75/127 (59%), Positives = 90/127 (70%), Gaps = 6/127 (4%)
Query 19 PEEEDVKDLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEPAYKKLV 78
PE V+D A+K VLK AL+HDGLARG+ E +KALD ++A C L ESC + AY KLV
Sbjct 24 PETVSVED---ALKEVLKRALVHDGLARGIREASKALDRRQAHLCVLCESCDQEAYVKLV 80
Query 79 EGLCKEHGIPLIEVADSKELGQWAGLCRIDNEGAARKVVGASCVAVTDFGEQSEALIFLQ 138
E LC E PLI+VAD K LG+WAGLC +D +G ARKVVG SCVAVTD+GE S A LQ
Sbjct 81 EALCAESETPLIKVADPKVLGEWAGLCVLDRDGNARKVVGCSCVAVTDYGEDSAA---LQ 137
Query 139 NHIKSLS 145
++S S
Sbjct 138 KLLESFS 144
> SPBC1685.02c
Length=148
Score = 137 bits (344), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 67/108 (62%), Positives = 81/108 (75%), Gaps = 0/108 (0%)
Query 30 AIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEPAYKKLVEGLCKEHGIPL 89
++K VLK AL+HDGLARG+ E +KALD ++A C L ESC + AY KLVE LC E PL
Sbjct 35 SLKEVLKRALVHDGLARGIREASKALDRRQAHLCVLCESCDQEAYVKLVEALCAESQTPL 94
Query 90 IEVADSKELGQWAGLCRIDNEGAARKVVGASCVAVTDFGEQSEALIFL 137
++VAD K LG+WAGLC +D +G ARKVVG SCVAVTD+GE S AL L
Sbjct 95 VKVADPKILGEWAGLCVLDRDGNARKVVGCSCVAVTDYGEDSVALQTL 142
> 7294509
Length=139
Score = 135 bits (341), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 65/127 (51%), Positives = 86/127 (67%), Gaps = 0/127 (0%)
Query 19 PEEEDVKDLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEPAYKKLV 78
P + D+ TA++ VLK +LI DGL G+H+ KALD ++A C LAES EP YKKLV
Sbjct 13 PVLDGAMDINTALQEVLKKSLIADGLVHGIHQACKALDKRQAVLCILAESFDEPNYKKLV 72
Query 79 EGLCKEHGIPLIEVADSKELGQWAGLCRIDNEGAARKVVGASCVAVTDFGEQSEALIFLQ 138
LC EH IPLI V K+LG+W+GLC+ID EG RKV G S V + DFGE++ AL ++
Sbjct 73 TALCNEHQIPLIRVDSHKKLGEWSGLCKIDKEGKPRKVCGCSVVVIKDFGEETPALDVVK 132
Query 139 NHIKSLS 145
+H++ S
Sbjct 133 DHLRQNS 139
> Hs18553260
Length=132
Score = 130 bits (326), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 64/119 (53%), Positives = 84/119 (70%), Gaps = 0/119 (0%)
Query 24 VKDLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEPAYKKLVEGLCK 83
V D+ TA++ VLK ALIH+GLA G+ + AKALD ++A C LA +C EP KLVE LC
Sbjct 11 VMDVNTALQEVLKTALIHNGLACGICKAAKALDKRQAHLCVLASNCDEPMDVKLVEALCA 70
Query 84 EHGIPLIEVADSKELGQWAGLCRIDNEGAARKVVGASCVAVTDFGEQSEALIFLQNHIK 142
EH I LI+V D+K+LG+W GLC+ D EG RKVVG SCV V D+G +S+A ++ + K
Sbjct 71 EHQIDLIKVDDNKKLGEWVGLCKTDREGKPRKVVGRSCVVVKDYGTESQAKDVIEEYFK 129
> At2g32060
Length=144
Score = 129 bits (324), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 60/125 (48%), Positives = 87/125 (69%), Gaps = 3/125 (2%)
Query 19 PEEEDVKDLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEPAYKKLV 78
PE+ DV TA++ ++ + + G+ RGLHE AK ++ + AQ C LAE C++P Y KLV
Sbjct 22 PEDMDVS---TALELTVRKSRAYGGVVRGLHESAKLIEKRNAQLCVLAEDCNQPDYVKLV 78
Query 79 EGLCKEHGIPLIEVADSKELGQWAGLCRIDNEGAARKVVGASCVAVTDFGEQSEALIFLQ 138
+ LC +H I L+ V +K LG+WAGLC+ID+EG ARKVVG SC+ + DFGE++ AL ++
Sbjct 79 KALCADHSIKLLTVPSAKTLGEWAGLCKIDSEGNARKVVGCSCLVIKDFGEETTALNIVK 138
Query 139 NHIKS 143
H+ S
Sbjct 139 KHLDS 143
> At1g15930
Length=144
Score = 127 bits (318), Expect = 9e-30, Method: Compositional matrix adjust.
Identities = 60/118 (50%), Positives = 82/118 (69%), Gaps = 0/118 (0%)
Query 26 DLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEPAYKKLVEGLCKEH 85
DL TA++ L+ A + G+ RGLHE AK ++ + AQ LAE C++P Y KLV+ LC +H
Sbjct 26 DLMTALELTLRKARAYGGVVRGLHECAKLIEKRVAQLVVLAEDCNQPDYVKLVKALCADH 85
Query 86 GIPLIEVADSKELGQWAGLCRIDNEGAARKVVGASCVAVTDFGEQSEALIFLQNHIKS 143
+ L+ V +K LG+WAGLC+ID+EG ARKVVG SC+ V DFGE++ AL + HI S
Sbjct 86 EVRLLTVPSAKTLGEWAGLCKIDSEGNARKVVGCSCLVVKDFGEETTALSIVNKHIAS 143
> CE26896
Length=140
Score = 123 bits (309), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 69/143 (48%), Positives = 89/143 (62%), Gaps = 7/143 (4%)
Query 3 MSDVESAGAPEEVQVEPEE--EDVKDLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRA 60
MSD A +VQV P + D + A++ VL+ A DGLA+GLHE KALD + A
Sbjct 1 MSD-----AGGDVQVAPAAVAQGPMDKEGALRAVLRAAHHADGLAKGLHETCKALDKREA 55
Query 61 QACFLAESCSEPAYKKLVEGLCKEHGIPLIEVADSKELGQWAGLCRIDNEGAARKVVGAS 120
C LAE+C EP Y KLVE LC EH IPLI+VAD K +G++ GLC+ D EG ARKVVG S
Sbjct 56 HFCVLAENCDEPQYVKLVETLCAEHQIPLIKVADKKIIGEYCGLCKYDKEGKARKVVGCS 115
Query 121 CVAVTDFGEQSEALIFLQNHIKS 143
VT++G + + L ++ S
Sbjct 116 SAVVTNWGNEEQGRAILTDYFAS 138
> Hs17463746
Length=118
Score = 105 bits (263), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 58/119 (48%), Positives = 76/119 (63%), Gaps = 14/119 (11%)
Query 24 VKDLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEPAYKKLVEGLCK 83
V D+ TA++ VLK ALIHDGLARG+HE A+ALD +A VE LC
Sbjct 11 VMDVNTALQEVLKTALIHDGLARGIHEAAEALDKCQAHL--------------FVEALCH 56
Query 84 EHGIPLIEVADSKELGQWAGLCRIDNEGAARKVVGASCVAVTDFGEQSEALIFLQNHIK 142
EH I LI+V D+K+LG+ GLC+I+ EG KVVG SCV V D+G++S+A +Q + K
Sbjct 57 EHQINLIKVDDNKKLGERIGLCKINREGKPCKVVGCSCVVVKDYGKESQAKDVIQEYFK 115
> YOR369c
Length=143
Score = 98.2 bits (243), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 59/117 (50%), Positives = 78/117 (66%), Gaps = 2/117 (1%)
Query 27 LKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEPAYKKLVEGLCK--E 84
++ A+K VL+ AL+HDGLARGL E KAL A L S +E KLVEGL E
Sbjct 24 IEDALKVVLRTALVHDGLARGLRESTKALTRGEALLVVLVSSVTEANIIKLVEGLANDPE 83
Query 85 HGIPLIEVADSKELGQWAGLCRIDNEGAARKVVGASCVAVTDFGEQSEALIFLQNHI 141
+ +PLI+VAD+K+LG+WAGL +ID EG ARKVVGAS V V ++G +++ L + H
Sbjct 84 NKVPLIKVADAKQLGEWAGLGKIDREGNARKVVGASVVVVKNWGAETDELSMIMEHF 140
> Hs20548341
Length=110
Score = 88.6 bits (218), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 41/78 (52%), Positives = 54/78 (69%), Gaps = 0/78 (0%)
Query 24 VKDLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEPAYKKLVEGLCK 83
V D+ TA++ VLK LIHDGLARG+HE AK LD +A LA +C E Y KLVE +C
Sbjct 11 VMDINTALQEVLKTTLIHDGLARGIHEAAKPLDKGQAHLYVLASNCDETVYVKLVEAICA 70
Query 84 EHGIPLIEVADSKELGQW 101
+H I I+V D+K++G+W
Sbjct 71 KHQINFIKVDDNKKVGEW 88
> Hs20471875
Length=115
Score = 78.2 bits (191), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 46/119 (38%), Positives = 64/119 (53%), Gaps = 17/119 (14%)
Query 24 VKDLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEPAYKKLVEGLCK 83
+ ++ TA++ VLK AL+HDG A G+ E AKALD +A C LA +C EP Y KLVE C
Sbjct 11 IMEVNTALQEVLKTALVHDGPACGILEAAKALDKCQAHLCVLASNCDEPVYVKLVEAFCA 70
Query 84 EHGIPLIEVADSKELGQWAGLCRIDNEGAARKVVGASCVAVTDFGEQSEALIFLQNHIK 142
EH ++ +S KVVG SCV V D G++ +A ++ + K
Sbjct 71 EHRTNRLKRGES-----------------GCKVVGGSCVEVKDAGKECQAKDVIKEYFK 112
> Hs17462075_1
Length=90
Score = 72.4 bits (176), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 40/76 (52%), Positives = 48/76 (63%), Gaps = 3/76 (3%)
Query 24 VKDLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFL---AESCSEPAYKKLVEG 80
V D+ T ++ VLK ALIHDGLA + + AKA D +A C L A +C EP Y KLVE
Sbjct 11 VMDINTVLQEVLKTALIHDGLAYEICKAAKASDKCQAHLCVLCVLASNCDEPMYVKLVEA 70
Query 81 LCKEHGIPLIEVADSK 96
LC EH I LI+V D K
Sbjct 71 LCAEHQINLIKVDDQK 86
> Hs22055843
Length=96
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 43/85 (50%), Positives = 51/85 (60%), Gaps = 4/85 (4%)
Query 24 VKDLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEPAYKKLVEGLCK 83
V DL TA++ VLK ALIHDGLARG+ E AKALD A C SC LV+ L
Sbjct 11 VMDLNTALQEVLKAALIHDGLARGIREAAKALDKCWAHPCAAPNSC---LCGWLVKALVA 67
Query 84 EHGIPLIEVADSKELGQWAGLCRID 108
EH I LI+V D+K LG+ C+ D
Sbjct 68 EHQITLIKVDDNK-LGEGMDHCKTD 91
> Hs22043188
Length=310
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 31/65 (47%), Positives = 40/65 (61%), Gaps = 4/65 (6%)
Query 69 CSEPAYKKLVEGLCKEHGIPLIEVADSKELGQWAGLCRIDNEGAARKVVGASCVAVTDFG 128
C EP Y LV LC EH I L +V D K+LG+ GL EG KV+G +CV V D+G
Sbjct 86 CDEPVYVMLVVALCAEHQIYLSKVDDRKKLGEEIGL----REGKPYKVIGCNCVVVKDYG 141
Query 129 EQSEA 133
++S+A
Sbjct 142 KESQA 146
> ECU01g0920
Length=134
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 39/122 (31%), Positives = 61/122 (50%), Gaps = 4/122 (3%)
Query 18 EPEEEDVKDLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEPAYKKL 77
EP E L+ A+ +V K + + L+RG E K + + + +AE+ +EP KL
Sbjct 6 EPMMEPEMTLQEALSKVCKVSRTYCKLSRGAKETTKKMLADKMSFVMVAEN-AEPRISKL 64
Query 78 VEGLCKEHGIPLIEVADSKELGQWAGLCRIDNEGAARKVVGASCVA-VTDFGEQSEALIF 136
V L K+ IP+I + ELG+ G+ + + G R CVA V D+ EQ+ F
Sbjct 65 VMALAKKKNIPVISIGSCLELGRIVGVENVSSSGKVRS--KGCCVAGVQDYCEQTSEAGF 122
Query 137 LQ 138
+Q
Sbjct 123 VQ 124
> At1g80800
Length=59
Score = 48.5 bits (114), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 24/51 (47%), Positives = 32/51 (62%), Gaps = 7/51 (13%)
Query 74 YKKLVEGLCKEHGIPLIEVADSKELGQWAGLCRIDNEGAARKVVGASCVAV 124
Y KLV+ LC +H I L+ + + LC+ID+EG ARKVVG SCV +
Sbjct 2 YVKLVKALCADHNINLLTLVN-------GPLCKIDSEGNARKVVGCSCVVI 45
> 7297543
Length=160
Score = 33.9 bits (76), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 36/134 (26%), Positives = 62/134 (46%), Gaps = 20/134 (14%)
Query 16 QVEPEEEDVKD--LKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEPA 73
+V P+ + D L I +L+ AL ++ L +G +E K L+ A LA +EP
Sbjct 37 EVNPKAFPLADAQLTAKIMNLLQQALNYNQLRKGANEATKTLNRGLADIVVLAGD-AEPI 95
Query 74 YKKL-VEGLCKEHGIPLIEVADSKELGQWAGLCRIDNEGAARKVVGASCVAVTDFGEQSE 132
L + LC++ +P + V + LG+ G+ +R +V +C T+ G Q
Sbjct 96 EILLHLPLLCEDKNVPYVFVRSKQALGRACGV--------SRPIV--ACSVTTNEGSQ-- 143
Query 133 ALIFLQNHIKSLSQ 146
L++ I S+ Q
Sbjct 144 ----LKSQITSIQQ 153
> At4g32350
Length=730
Score = 32.7 bits (73), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 37/75 (49%), Gaps = 2/75 (2%)
Query 20 EEEDVKDLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEPAYKKLVE 79
EE + + +K+ + L + +A+ HE A+ALD+ +A+ F E + P K V
Sbjct 503 EERETERMKSPFYKSLPPPYVKKSIAKARHEKAEALDNPKAR--FDGEEGNHPDNGKNVY 560
Query 80 GLCKEHGIPLIEVAD 94
G + +G EV D
Sbjct 561 GAERRNGAGHHEVND 575
> 7293712
Length=486
Score = 32.3 bits (72), Expect = 0.29, Method: Composition-based stats.
Identities = 26/75 (34%), Positives = 39/75 (52%), Gaps = 6/75 (8%)
Query 13 EEVQVEPEEEDVKDLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEP 72
E V+V EE+ + ++ A K VLKN L+ L + E L+S R + FLA +C+
Sbjct 381 EVVKVIQEEKLLANVDVAGK-VLKNGLL--SLEK---EFPHILNSTRGRGTFLAVNCTNT 434
Query 73 AYKKLVEGLCKEHGI 87
+ + G K HGI
Sbjct 435 KVRDQIIGALKLHGI 449
> CE19186
Length=163
Score = 32.3 bits (72), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 37/83 (44%), Gaps = 1/83 (1%)
Query 25 KDLKTAIKRVLKNALIHDGLAR-GLHEVAKALDSKRAQACFLAESCSEPAYKKLVEGLCK 83
+ L + +++K A D R G+ +V K L C LA + S + G+C+
Sbjct 49 RKLAKKVYKLIKKASAGDKTLREGIKDVQKELRRNEKGICILAGNVSPIDVYSHIPGICE 108
Query 84 EHGIPLIEVADSKELGQWAGLCR 106
E IP + + ++LG G R
Sbjct 109 EKEIPYVYIPSREQLGLAVGHRR 131
> YDL208w
Length=173
Score = 32.3 bits (72), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 34/82 (41%), Gaps = 0/82 (0%)
Query 25 KDLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEPAYKKLVEGLCKE 84
K L + + +K A + RG+ EV KAL +A S + LC++
Sbjct 54 KKLNKKVLKTVKKASKAKNVKRGVKEVVKALRKGEKGLVVIAGDISPADVISHIPVLCED 113
Query 85 HGIPLIEVADSKELGQWAGLCR 106
H +P I + ++LG R
Sbjct 114 HSVPYIFIPSKQDLGAAGATKR 135
> SPCC306.04c
Length=920
Score = 30.4 bits (67), Expect = 1.2, Method: Composition-based stats.
Identities = 18/61 (29%), Positives = 29/61 (47%), Gaps = 0/61 (0%)
Query 12 PEEVQVEPEEEDVKDLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSE 71
PEE E ++DL+ A+ R +K+ +I + + LH + K + Q L S S
Sbjct 367 PEEAYAEATSVVLRDLEAALLRDVKSKIIGPAIFKYLHSMPKPSVKEELQENLLVSSTSV 426
Query 72 P 72
P
Sbjct 427 P 427
> At5g08180
Length=156
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 38/82 (46%), Gaps = 0/82 (0%)
Query 25 KDLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEPAYKKLVEGLCKE 84
K L+ ++++ A L RG+ EV K++ + C +A + S + LC+E
Sbjct 32 KKLQKRTFKLIQKAAGKKCLKRGVKEVVKSIRRGQKGLCVIAGNISPIDVITHLPILCEE 91
Query 85 HGIPLIEVADSKELGQWAGLCR 106
G+P + V ++L Q R
Sbjct 92 AGVPYVYVPSKEDLAQAGATKR 113
> 7290722
Length=271
Score = 29.6 bits (65), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 25/95 (26%), Positives = 38/95 (40%), Gaps = 11/95 (11%)
Query 47 GLHEVAKALDSKRAQACFLAESCSEPAYKKLVEGLCKEHGIPLIEVADSKELGQWA---- 102
G + V K ++ K+AQ +A + LC++ G+P V LG+
Sbjct 144 GTNTVTKLIEQKKAQLVVIAHDVDPLELVLFLPALCRKMGVPYCIVKGKARLGRLVRRKT 203
Query 103 ----GLCRIDNEGAARKVVGASCVAV-TDFGEQSE 132
L +DN A G AV T+F E+ E
Sbjct 204 CTTLALTTVDNNDKAN--FGKVLEAVKTNFNERHE 236
> Hs8922444
Length=1045
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 34/142 (23%), Positives = 61/142 (42%), Gaps = 9/142 (6%)
Query 5 DVESAGAPEEVQVEPEEEDVKDLKTAIKRVLKNA--LIHDGLARGLHEVAKALDSKRAQA 62
D+ES+ E+V + + T ++ L+N+ + +DG R ++ A LDS
Sbjct 8 DIESSSREEDVSITSNLSSIDSFYTMVQDQLRNSYQIGYDGSLRIIY--ASGLDSHYQTE 65
Query 63 CFLAESCSEPAYKKLVEGLCKEHGIPLIEVADSKELGQWAGLCRIDNEGAARKVVGASCV 122
+ + P K L E+G L+E KE Q +++ G +V G + +
Sbjct 66 PHVLAGTANPTVAKRNMTLPGENGQNLVEWRFRKEQAQ----GKVNVFGRKLRVNGRNLL 121
Query 123 AVTDFGEQSEALIFLQNHIKSL 144
+V DF ++ +H K L
Sbjct 122 SV-DFDRTTKTEKIYDDHRKFL 142
> At1g53800
Length=603
Score = 29.3 bits (64), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 26/46 (56%), Gaps = 1/46 (2%)
Query 45 ARGLHEVAKALDSKRAQACFLAESCSEPAYKKLV-EGLCKEHGIPL 89
A+ K+ + +R A +A ++P+Y++ V GL K HGIP+
Sbjct 275 AKSNRRAPKSPEQRRRIAEAIAAKWADPSYRERVCSGLAKYHGIPV 320
> SPBC29A3.04
Length=259
Score = 28.5 bits (62), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 14/53 (26%), Positives = 25/53 (47%), Gaps = 0/53 (0%)
Query 47 GLHEVAKALDSKRAQACFLAESCSEPAYKKLVEGLCKEHGIPLIEVADSKELG 99
GL+ V +++K+A+ +A + LCK+ G+P V + LG
Sbjct 135 GLNHVVALIEAKKAKLVLIASDVDPIELVVFLPALCKKMGVPYAIVKNKARLG 187
> At2g43090
Length=251
Score = 27.7 bits (60), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 53 KALDSKRAQACFLAESCSEPAYKKLVEGLC 82
K L S+ A + F+ S +EP +K GLC
Sbjct 45 KPLVSREASSSFVTRSAAEPQERKTFHGLC 74
> At2g01740
Length=559
Score = 27.7 bits (60), Expect = 8.6, Method: Composition-based stats.
Identities = 26/101 (25%), Positives = 36/101 (35%), Gaps = 29/101 (28%)
Query 21 EEDVKDLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEPAYKKLVEG 80
E DV L T I + KN +H+ + F E ++ Y L++
Sbjct 370 EPDVVALSTMIDGIAKNGQLHEAIVY-----------------FCIEKANDVMYTVLIDA 412
Query 81 LCKEHGIPLIE-----------VADSKELGQW-AGLCRIDN 109
LCKE +E V D W AGLC+ N
Sbjct 413 LCKEGDFIEVERLFSKISEAGLVPDKFMYTSWIAGLCKQGN 453
> SPAC1782.10c
Length=154
Score = 27.3 bits (59), Expect = 10.0, Method: Compositional matrix adjust.
Identities = 23/108 (21%), Positives = 42/108 (38%), Gaps = 11/108 (10%)
Query 25 KDLKTAIKRVLKNALIHDGLARGLHEVAKALDSKRAQACFLAESCSEPAYKKLVEGLCKE 84
K L + + +K A + RG+ EV KA+ LA S + LC++
Sbjct 35 KKLNKKMMKTVKKASKQKHILRGVKEVVKAVRKGEKGLVILAGDISPMDVISHIPVLCED 94
Query 85 HGIPLIEVADSKELGQWAGLCRIDNEGAARKVVGASCVAVTDFGEQSE 132
+ +P + + LG+ + R SCV + G++ +
Sbjct 95 NNVPYLYTVSKELLGEASNTKR-----------PTSCVMIVPGGKKKD 131
Lambda K H
0.314 0.131 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1748847648
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40