bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2188_orf1
Length=137
Score E
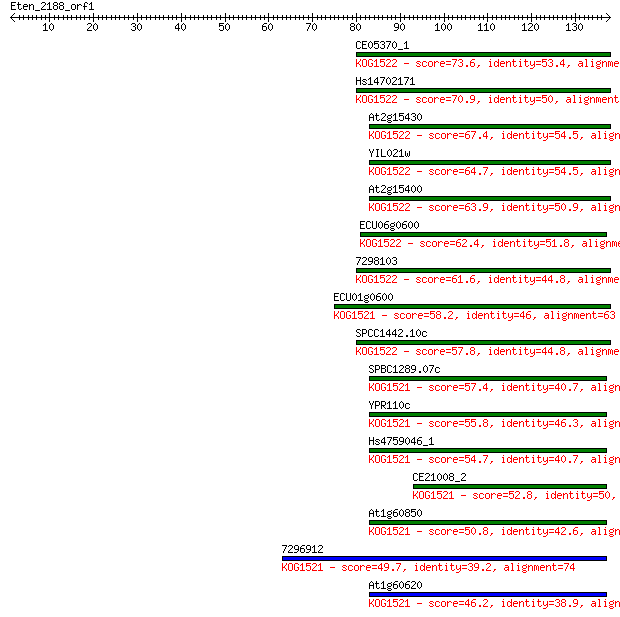
Sequences producing significant alignments: (Bits) Value
CE05370_1 73.6 1e-13
Hs14702171 70.9 6e-13
At2g15430 67.4 7e-12
YIL021w 64.7 5e-11
At2g15400 63.9 9e-11
ECU06g0600 62.4 3e-10
7298103 61.6 4e-10
ECU01g0600 58.2 5e-09
SPCC1442.10c 57.8 5e-09
SPBC1289.07c 57.4 7e-09
YPR110c 55.8 2e-08
Hs4759046_1 54.7 5e-08
CE21008_2 52.8 2e-07
At1g60850 50.8 8e-07
7296912 49.7 2e-06
At1g60620 46.2 2e-05
> CE05370_1
Length=303
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 31/58 (53%), Positives = 44/58 (75%), Gaps = 0/58 (0%)
Query 80 QAAIRVTELSNKVVRFTLNNCDAAIANALRRVMISEVPTLAIDLATVFENTSVLHDEY 137
Q I VTEL+N +++F L + D ++AN+LRRV ++EVPT+AID + NTSVLHDE+
Sbjct 6 QPNIEVTELTNDIIKFVLWDTDLSVANSLRRVFMAEVPTIAIDWVQIETNTSVLHDEF 63
> Hs14702171
Length=275
Score = 70.9 bits (172), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 29/58 (50%), Positives = 44/58 (75%), Gaps = 0/58 (0%)
Query 80 QAAIRVTELSNKVVRFTLNNCDAAIANALRRVMISEVPTLAIDLATVFENTSVLHDEY 137
Q +R+TEL+++ V+F + N D A+AN++RRV I+EVP +AID + N+SVLHDE+
Sbjct 6 QPTVRITELTDENVKFIIENTDLAVANSIRRVFIAEVPIIAIDWVQIDANSSVLHDEF 63
> At2g15430
Length=319
Score = 67.4 bits (163), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 30/55 (54%), Positives = 41/55 (74%), Gaps = 0/55 (0%)
Query 83 IRVTELSNKVVRFTLNNCDAAIANALRRVMISEVPTLAIDLATVFENTSVLHDEY 137
I++ EL + +F L D ++ANALRRVMISEVPT+AIDL + N+SVL+DE+
Sbjct 12 IKIRELKDDYAKFELRETDVSMANALRRVMISEVPTVAIDLVEIEVNSSVLNDEF 66
> YIL021w
Length=318
Score = 64.7 bits (156), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 30/55 (54%), Positives = 40/55 (72%), Gaps = 0/55 (0%)
Query 83 IRVTELSNKVVRFTLNNCDAAIANALRRVMISEVPTLAIDLATVFENTSVLHDEY 137
+++ E S V F L+N D A+AN+LRRVMI+E+PTLAID V NT+VL DE+
Sbjct 8 VKIREASKDNVDFILSNVDLAMANSLRRVMIAEIPTLAIDSVEVETNTTVLADEF 62
> At2g15400
Length=319
Score = 63.9 bits (154), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 28/55 (50%), Positives = 40/55 (72%), Gaps = 0/55 (0%)
Query 83 IRVTELSNKVVRFTLNNCDAAIANALRRVMISEVPTLAIDLATVFENTSVLHDEY 137
+++ EL + +F L D ++ANALRRVMISEVPT+AI L + N+SVL+DE+
Sbjct 12 VKIRELKDDYAKFELRETDVSMANALRRVMISEVPTMAIHLVKIEVNSSVLNDEF 66
> ECU06g0600
Length=240
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 29/56 (51%), Positives = 40/56 (71%), Gaps = 0/56 (0%)
Query 81 AAIRVTELSNKVVRFTLNNCDAAIANALRRVMISEVPTLAIDLATVFENTSVLHDE 136
A+ V EL+ + +RFTL+N ANALRR++ISEVPT+AID+ + N +VL DE
Sbjct 2 VALTVEELTPEHIRFTLSNTTVGYANALRRILISEVPTIAIDMVEIERNNTVLPDE 57
> 7298103
Length=275
Score = 61.6 bits (148), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 26/58 (44%), Positives = 43/58 (74%), Gaps = 0/58 (0%)
Query 80 QAAIRVTELSNKVVRFTLNNCDAAIANALRRVMISEVPTLAIDLATVFENTSVLHDEY 137
Q ++++TEL++ V+F L + + ++AN+LRRV I+E PTLAID + N++VL DE+
Sbjct 6 QPSVQITELTDDNVKFVLEDTELSVANSLRRVFIAETPTLAIDWVQLEANSTVLSDEF 63
> ECU01g0600
Length=305
Score = 58.2 bits (139), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 29/63 (46%), Positives = 43/63 (68%), Gaps = 0/63 (0%)
Query 75 QQQLMQAAIRVTELSNKVVRFTLNNCDAAIANALRRVMISEVPTLAIDLATVFENTSVLH 134
+Q + + IR+TE S++ + F L N D +IANALRR++ SEVPT+A+ + EN SV
Sbjct 24 EQTVDEIEIRITEYSSEKLEFDLINVDCSIANALRRILSSEVPTMAVRDIFLKENESVFP 83
Query 135 DEY 137
DE+
Sbjct 84 DEF 86
> SPCC1442.10c
Length=297
Score = 57.8 bits (138), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 26/58 (44%), Positives = 39/58 (67%), Gaps = 0/58 (0%)
Query 80 QAAIRVTELSNKVVRFTLNNCDAAIANALRRVMISEVPTLAIDLATVFENTSVLHDEY 137
+ I + +S V F L N A+AN+LRRV+++E+PT+AIDL + NTSV+ DE+
Sbjct 4 ETHITIRNISKNSVDFVLTNTSLAVANSLRRVVLAEIPTVAIDLVEINVNTSVMPDEF 61
> SPBC1289.07c
Length=348
Score = 57.4 bits (137), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 22/54 (40%), Positives = 37/54 (68%), Gaps = 0/54 (0%)
Query 83 IRVTELSNKVVRFTLNNCDAAIANALRRVMISEVPTLAIDLATVFENTSVLHDE 136
+ +T L + + F ++ DA+IANA RR++I+E+PTLA + + NTS++ DE
Sbjct 48 VSITSLDQETMVFEISGIDASIANAFRRILIAEIPTLAFEFVYIINNTSIIQDE 101
> YPR110c
Length=335
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 25/54 (46%), Positives = 35/54 (64%), Gaps = 0/54 (0%)
Query 83 IRVTELSNKVVRFTLNNCDAAIANALRRVMISEVPTLAIDLATVFENTSVLHDE 136
+ ++ L + F L N D +IANA RR+MISEVP++A + F NTSV+ DE
Sbjct 42 VNISSLDAREANFDLINIDTSIANAFRRIMISEVPSVAAEYVYFFNNTSVIQDE 95
> Hs4759046_1
Length=314
Score = 54.7 bits (130), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 22/54 (40%), Positives = 36/54 (66%), Gaps = 0/54 (0%)
Query 83 IRVTELSNKVVRFTLNNCDAAIANALRRVMISEVPTLAIDLATVFENTSVLHDE 136
+ V + + F + DAAIANA RR++++EVPT+A++ V+ NTS++ DE
Sbjct 51 VDVVHMDENSLEFDMVGIDAAIANAFRRILLAEVPTMAVEKVLVYNNTSIVQDE 104
> CE21008_2
Length=362
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 22/44 (50%), Positives = 34/44 (77%), Gaps = 0/44 (0%)
Query 93 VRFTLNNCDAAIANALRRVMISEVPTLAIDLATVFENTSVLHDE 136
+ F + +A IANALRRV+I+EVPT+A++ +++NTSV+ DE
Sbjct 64 LEFDITRIEAPIANALRRVLIAEVPTMALEKIYLYQNTSVIQDE 107
> At1g60850
Length=375
Score = 50.8 bits (120), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 35/54 (64%), Gaps = 0/54 (0%)
Query 83 IRVTELSNKVVRFTLNNCDAAIANALRRVMISEVPTLAIDLATVFENTSVLHDE 136
+ V L+ + F + DAA ANA RR++I+EVP++AI+ + NTSV+ DE
Sbjct 76 VDVVSLTKTDMEFDMIGIDAAFANAFRRILIAEVPSMAIEKVLIAYNTSVIIDE 129
> 7296912
Length=333
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 44/81 (54%), Gaps = 8/81 (9%)
Query 63 RLQQQPQLQQQQQQQLMQAAIRVTELSNKVVR-------FTLNNCDAAIANALRRVMISE 115
R++Q PQ A + +L K+VR F L AIANA RR+M+S+
Sbjct 17 RVKQDPQDYGLADDVFSVQAFK-EKLHIKIVRNDEDGLEFDLIGVYPAIANAFRRLMLSD 75
Query 116 VPTLAIDLATVFENTSVLHDE 136
VP++AI+ ++ NTS++ DE
Sbjct 76 VPSMAIEKVYIYNNTSIIQDE 96
> At1g60620
Length=385
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 34/54 (62%), Gaps = 0/54 (0%)
Query 83 IRVTELSNKVVRFTLNNCDAAIANALRRVMISEVPTLAIDLATVFENTSVLHDE 136
+ V L+ + F + A IANA RR++++E+P++AI+ V NTSV+ DE
Sbjct 83 VDVISLTETDMVFDMIGVHAGIANAFRRILLAELPSMAIEKVYVANNTSVIQDE 136
Lambda K H
0.322 0.127 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1461889840
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40