bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2198_orf1
Length=117
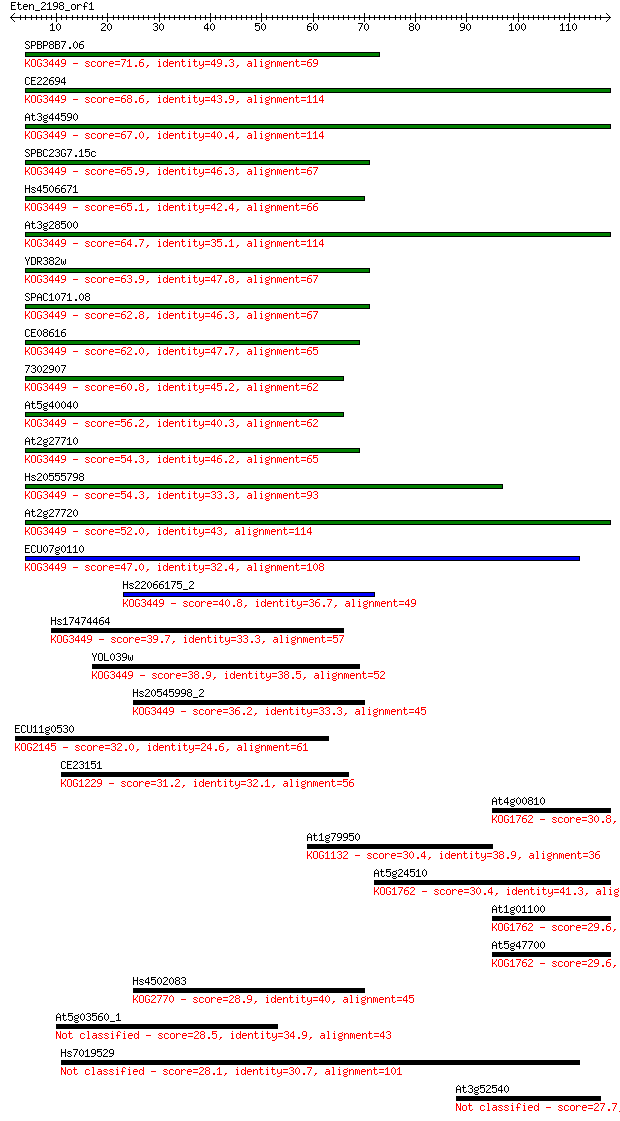
Score E
Sequences producing significant alignments: (Bits) Value
SPBP8B7.06 71.6 3e-13
CE22694 68.6 3e-12
At3g44590 67.0 8e-12
SPBC23G7.15c 65.9 2e-11
Hs4506671 65.1 3e-11
At3g28500 64.7 4e-11
YDR382w 63.9 7e-11
SPAC1071.08 62.8 1e-10
CE08616 62.0 2e-10
7302907 60.8 5e-10
At5g40040 56.2 1e-08
At2g27710 54.3 5e-08
Hs20555798 54.3 5e-08
At2g27720 52.0 3e-07
ECU07g0110 47.0 8e-06
Hs22066175_2 40.8 6e-04
Hs17474464 39.7 0.001
YOL039w 38.9 0.003
Hs20545998_2 36.2 0.016
ECU11g0530 32.0 0.28
CE23151 31.2 0.49
At4g00810 30.8 0.55
At1g79950 30.4 0.82
At5g24510 30.4 0.90
At1g01100 29.6 1.5
At5g47700 29.6 1.6
Hs4502083 28.9 2.5
At5g03560_1 28.5 3.3
Hs7019529 28.1 4.6
At3g52540 27.7 5.2
> SPBP8B7.06
Length=110
Score = 71.6 bits (174), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 34/69 (49%), Positives = 48/69 (69%), Gaps = 0/69 (0%)
Query 4 MKYVAAYLMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGLE 63
MKY+AAYL+ +GG ++P+A+D+ VL VG E E ++TLI+ + GK E+I+AG E
Sbjct 1 MKYLAAYLLLTVGGKDSPSASDIESVLSTVGIEAESERIETLINELNGKDIDELIAAGNE 60
Query 64 KLQKVPCGG 72
KL VP GG
Sbjct 61 KLATVPTGG 69
> CE22694
Length=110
Score = 68.6 bits (166), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 50/114 (43%), Positives = 64/114 (56%), Gaps = 4/114 (3%)
Query 4 MKYVAAYLMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGLE 63
M+YV+AYL+ VLGGN P D+ +L AVG + + E K ++ + GK E+I+ G
Sbjct 1 MRYVSAYLLAVLGGNANPKVDDLKNILSAVGVDADAETAKLVVSRLAGKTVEELIAEGSA 60
Query 64 KLQKVPCGGGAAAAAPAAAAAAGGGDSSSAAKETKKEEPEEEEEDGDMGLSLFD 117
L V G AAAA AA A S K KKEEP+EE +D DMG LFD
Sbjct 61 GLVSVSGGAAPAAAAAPAAGGAAPAADS---KPAKKEEPKEESDD-DMGFGLFD 110
> At3g44590
Length=111
Score = 67.0 bits (162), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 46/114 (40%), Positives = 75/114 (65%), Gaps = 3/114 (2%)
Query 4 MKYVAAYLMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGLE 63
MK AA+L+ VLGGN P+A ++ ++ AVGA+V+ E ++ L+ + GK E+I++G E
Sbjct 1 MKVAAAFLLAVLGGNANPSADNIKDIIGAVGADVDGESIELLLKEVSGKDIAELIASGRE 60
Query 64 KLQKVPCGGGAAAAAPAAAAAAGGGDSSSAAKETKKEEPEEEEEDGDMGLSLFD 117
KL VP GGG A +A ++ G +++ A++ + ++ E+EE D DMG SLF+
Sbjct 61 KLASVPSGGGVAVSAAPSSGGGG---AAAPAEKKEAKKEEKEESDDDMGFSLFE 111
> SPBC23G7.15c
Length=110
Score = 65.9 bits (159), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 31/67 (46%), Positives = 45/67 (67%), Gaps = 0/67 (0%)
Query 4 MKYVAAYLMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGLE 63
MKY+AAYL+ +GG ++P+A+D+ VL VG E E +++LI + GK E+I+AG E
Sbjct 1 MKYLAAYLLLTVGGKQSPSASDIESVLSTVGIEAEAERVESLISELNGKNIEELIAAGNE 60
Query 64 KLQKVPC 70
KL VP
Sbjct 61 KLSTVPS 67
> Hs4506671
Length=115
Score = 65.1 bits (157), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 28/66 (42%), Positives = 45/66 (68%), Gaps = 0/66 (0%)
Query 4 MKYVAAYLMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGLE 63
M+YVA+YL+ LGGN +P+A D+ ++L++VG E + + L +I + GK +VI+ G+
Sbjct 1 MRYVASYLLAALGGNSSPSAKDIKKILDSVGIEADDDRLNKVISELNGKNIEDVIAQGIG 60
Query 64 KLQKVP 69
KL VP
Sbjct 61 KLASVP 66
> At3g28500
Length=115
Score = 64.7 bits (156), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 40/115 (34%), Positives = 62/115 (53%), Gaps = 1/115 (0%)
Query 4 MKYVAAYLMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGLE 63
MK +AA+L+ LGGNE PT+ D+ ++LE+VGAE++ + L ++ E+I+AG E
Sbjct 1 MKVIAAFLLAKLGGNENPTSNDLKKILESVGAEIDETKIDLLFSLIKDHDVTELIAAGRE 60
Query 64 KLQKVPCGGGAAAAAPAAAAAAGGGDSSSAAKETKKEEPEEEEEDGDMG-LSLFD 117
K+ + GG A A + A+ KK E ++E D G + LFD
Sbjct 61 KMSALSSGGPAVAMVAGGGGGGAASAAEPVAESKKKVEEVKDESSDDAGMMGLFD 115
> YDR382w
Length=110
Score = 63.9 bits (154), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 32/68 (47%), Positives = 48/68 (70%), Gaps = 1/68 (1%)
Query 4 MKYVAAYLMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGKAA-HEVISAGL 62
MKY+AAYL+ V GGN AP+AAD+ V+E+VGAEV+ + L+ +++GK + E+I+ G
Sbjct 1 MKYLAAYLLLVQGGNAAPSAADIKAVVESVGAEVDEARINELLSSLEGKGSLEEIIAEGQ 60
Query 63 EKLQKVPC 70
+K VP
Sbjct 61 KKFATVPT 68
> SPAC1071.08
Length=110
Score = 62.8 bits (151), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 31/67 (46%), Positives = 43/67 (64%), Gaps = 0/67 (0%)
Query 4 MKYVAAYLMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGLE 63
MKY+AAYL+ +GG +P+A+D+ VL VG E E ++ LI + GK E+I+AG E
Sbjct 1 MKYLAAYLLLTVGGKNSPSASDIESVLSTVGIESESERVEALIKELDGKDIDELIAAGNE 60
Query 64 KLQKVPC 70
KL VP
Sbjct 61 KLATVPS 67
> CE08616
Length=136
Score = 62.0 bits (149), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 31/65 (47%), Positives = 44/65 (67%), Gaps = 0/65 (0%)
Query 4 MKYVAAYLMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGLE 63
MKY+ AYL+ LGGN +P+A DV +VLEA G + + E +++DA++GK EVI+ G
Sbjct 1 MKYLGAYLLATLGGNASPSAQDVLKVLEAGGLDCDMENANSVVDALKGKTISEVIAQGKV 60
Query 64 KLQKV 68
KL V
Sbjct 61 KLSSV 65
> 7302907
Length=113
Score = 60.8 bits (146), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 28/62 (45%), Positives = 43/62 (69%), Gaps = 0/62 (0%)
Query 4 MKYVAAYLMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGLE 63
M+YVAAYL+ VLGG ++P +D+ ++L +VG EV+ E L +I + GK+ ++I G E
Sbjct 1 MRYVAAYLLAVLGGKDSPANSDLEKILSSVGVEVDAERLTKVIKELAGKSIDDLIKEGRE 60
Query 64 KL 65
KL
Sbjct 61 KL 62
> At5g40040
Length=114
Score = 56.2 bits (134), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 41/62 (66%), Gaps = 0/62 (0%)
Query 4 MKYVAAYLMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGLE 63
MK VAAYL+ L GNE P+ AD+ +++E+VGAE++ E + ++ + E+I+ G E
Sbjct 1 MKVVAAYLLAKLSGNENPSVADLKKIVESVGAEIDQEKIDLFFSLIKDRDVTELIAVGRE 60
Query 64 KL 65
K+
Sbjct 61 KM 62
> At2g27710
Length=115
Score = 54.3 bits (129), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 30/65 (46%), Positives = 43/65 (66%), Gaps = 0/65 (0%)
Query 4 MKYVAAYLMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGLE 63
MK VAAYL+ VL G +PT+AD+ +L +VGAE ++ L+ ++GK E+I+AG E
Sbjct 1 MKVVAAYLLAVLSGKASPTSADIKTILGSVGAETEDSQIELLLKEVKGKDLAELIAAGRE 60
Query 64 KLQKV 68
KL V
Sbjct 61 KLASV 65
> Hs20555798
Length=93
Score = 54.3 bits (129), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 31/93 (33%), Positives = 52/93 (55%), Gaps = 1/93 (1%)
Query 4 MKYVAAYLMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGLE 63
M Y+ +YL+ L N + +A D+ ++L+ VG E + L +I + GK ++I+ G+
Sbjct 1 MVYLTSYLLPSLMSNTSLSAKDIKKILDRVGMEATDDWLNKVISELNGKNIEDIIAQGIG 60
Query 64 KLQKVPCGGGAAAAAPAAAAAAGGGDSSSAAKE 96
+L VP GG A +A +A G S+ AA+E
Sbjct 61 ELASVPAGGAVALSASLGSAGPAAG-STPAAEE 92
> At2g27720
Length=115
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 49/115 (42%), Positives = 72/115 (62%), Gaps = 1/115 (0%)
Query 4 MKYVAAYLMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGLE 63
MK VAA+L+ VL G +PT D+ +L +VGAE ++ L+ ++GK E+I+AG E
Sbjct 1 MKVVAAFLLAVLSGKASPTTGDIKDILGSVGAETEDSQIELLLKEVKGKDLAELIAAGRE 60
Query 64 KLQKVPCGGGAAAAAPAAAAAAGGGDSSSAA-KETKKEEPEEEEEDGDMGLSLFD 117
KL VP GGG A +A + GGG + AA + ++++ E+EE D DMG SLF+
Sbjct 61 KLASVPSGGGGGVAVASATSGGGGGGGAPAAESKKEEKKEEKEESDDDMGFSLFE 115
> ECU07g0110
Length=103
Score = 47.0 bits (110), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 35/110 (31%), Positives = 55/110 (50%), Gaps = 13/110 (11%)
Query 4 MKYVAAYLMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGLE 63
M+YVAAY+M G E + ++ + +GAE+ PE ++ + + GK+ EV+S G E
Sbjct 1 MEYVAAYVMFDKVGKELNERS-MTELFNEIGAEIEPETMRLFLSKVSGKSMDEVMSKGKE 59
Query 64 KLQKVPCGGGAAA--AAPAAAAAAGGGDSSSAAKETKKEEPEEEEEDGDM 111
+ + + A PA A S+ A E K EEE+ED D+
Sbjct 60 LMASLAISSSQKSEPAQPADTA------ESTQATENK----EEEDEDFDI 99
> Hs22066175_2
Length=72
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 31/49 (63%), Gaps = 0/49 (0%)
Query 23 AADVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGLEKLQKVPCG 71
A D+ R+L++VG E + + L +I + GK+ +VI+ G+ K+ VP G
Sbjct 2 AKDIKRILDSVGIEADDDRLNEVISELNGKSIEDVIAQGIGKVASVPAG 50
> Hs17474464
Length=116
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 30/57 (52%), Gaps = 0/57 (0%)
Query 9 AYLMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGLEKL 65
+YL+ LGGN P+ DV ++L + + N +I + GK VI+ G+ KL
Sbjct 28 SYLLAALGGNPLPSTKDVKKILYNISNKANGNQCSKIISELNGKNIKGVIAQGIGKL 84
> YOL039w
Length=106
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 33/52 (63%), Gaps = 1/52 (1%)
Query 17 GNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGLEKLQKV 68
GN P A + +LE+VG E+ E + +++ A++GK+ E+I+ G EKL V
Sbjct 14 GN-TPDATKIKAILESVGIEIEDEKVSSVLSALEGKSVDELITEGNEKLAAV 64
> Hs20545998_2
Length=76
Score = 36.2 bits (82), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 27/45 (60%), Gaps = 0/45 (0%)
Query 25 DVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGLEKLQKVP 69
D+ ++L++VG E + + L +I + GK +VI+ + KL VP
Sbjct 2 DIKKILDSVGIETDNDRLNKVISELNGKNIEDVIAKSISKLANVP 46
> ECU11g0530
Length=385
Score = 32.0 bits (71), Expect = 0.28, Method: Composition-based stats.
Identities = 15/61 (24%), Positives = 29/61 (47%), Gaps = 0/61 (0%)
Query 2 MAMKYVAAYLMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAG 61
MA + + + + V+ +E P A D +++ G E + L ++ + GK AH G
Sbjct 1 MAEQRITPWDVEVVSTDEVPVAIDYDKIINQFGCEKFNQALADRLEKLSGKPAHYFFRRG 60
Query 62 L 62
+
Sbjct 61 I 61
> CE23151
Length=760
Score = 31.2 bits (69), Expect = 0.49, Method: Composition-based stats.
Identities = 18/56 (32%), Positives = 33/56 (58%), Gaps = 1/56 (1%)
Query 11 LMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGLEKLQ 66
L + GG EAP + +V +L V A V+ E +T+ DAM+ ++HE+ + + + +
Sbjct 332 LKSQTGGIEAPIS-EVLTMLRDVSARVDGEPAQTIKDAMKVLSSHELYAPSINRFR 386
> At4g00810
Length=110
Score = 30.8 bits (68), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 15/23 (65%), Positives = 17/23 (73%), Gaps = 1/23 (4%)
Query 95 KETKKEEPEEEEEDGDMGLSLFD 117
KE KK+EP EE DGD+G LFD
Sbjct 89 KEEKKDEPAEES-DGDLGFGLFD 110
> At1g79950
Length=983
Score = 30.4 bits (67), Expect = 0.82, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 59 SAGLEKLQKVPCGGGAAAAAPAAAAAAGGGDSSSAA 94
+ GL+K +KVP G+A+++ A GGGD A+
Sbjct 861 TMGLKKKRKVPESQGSASSSVLTAKGNGGGDKKEAS 896
> At5g24510
Length=111
Score = 30.4 bits (67), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 23/47 (48%), Gaps = 1/47 (2%)
Query 72 GGAAAAAPAAAAAAGGGDSSSAAKETKKE-EPEEEEEDGDMGLSLFD 117
GG A P AA S S +E K E E +EE + DM + LFD
Sbjct 65 GGCGVARPVTTAAPTASQSVSIPEEKKNEMEVIKEESEDDMIIGLFD 111
> At1g01100
Length=112
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 14/23 (60%), Positives = 17/23 (73%), Gaps = 1/23 (4%)
Query 95 KETKKEEPEEEEEDGDMGLSLFD 117
+E KK+EP EE DGD+G LFD
Sbjct 91 EEKKKDEPAEES-DGDLGFGLFD 112
> At5g47700
Length=113
Score = 29.6 bits (65), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 14/23 (60%), Positives = 17/23 (73%), Gaps = 1/23 (4%)
Query 95 KETKKEEPEEEEEDGDMGLSLFD 117
+E KK+EP EE DGD+G LFD
Sbjct 92 EEKKKDEPAEES-DGDLGFGLFD 113
> Hs4502083
Length=403
Score = 28.9 bits (63), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 28/46 (60%), Gaps = 1/46 (2%)
Query 25 DVSRVLEAVGAEVNPEVLKTLIDAMQGKAAHEVISAGL-EKLQKVP 69
D R L+ G +V EVL + A+QG A +V+ AG+ + L+K+P
Sbjct 158 DKVRELQNQGRDVGLEVLDNALLALQGPTAAQVLQAGVADDLRKLP 203
> At5g03560_1
Length=520
Score = 28.5 bits (62), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 23/43 (53%), Gaps = 0/43 (0%)
Query 10 YLMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGK 52
YL+ ++G +P AA + V EA E E + L+ M+GK
Sbjct 271 YLLEMMGNGMSPNAATYTAVFEAFVREGKEESARELLQEMKGK 313
> Hs7019529
Length=504
Score = 28.1 bits (61), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 31/109 (28%), Positives = 45/109 (41%), Gaps = 8/109 (7%)
Query 11 LMTVLGGNEAPTAADVSRVLEAVGAEVNPEVLKTLIDAMQGK------AAHEVISAGLEK 64
LM +G P+A + ++EA + P L+DA++ A EV AG+
Sbjct 368 LMAAVGAPRFPSALGLVLLVEAAAVLIGPPSAGRLVDALKNYEIIFYLAGSEVALAGVFM 427
Query 65 LQKVPCGGGAAAAAPAAAAAAGG-GDSSSAAKETKKEE-PEEEEEDGDM 111
C A AAP+ GG D+ A E E P EE G++
Sbjct 428 AVATNCCLRCAKAAPSGPGTEGGASDTEDAEAEGDSEPLPVVAEEPGNL 476
> At3g52540
Length=282
Score = 27.7 bits (60), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 15/28 (53%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 88 GDSSSAAKETKKEEPEEEEEDGDMGLSL 115
G S+S +E K + EEEEDG M LSL
Sbjct 114 GTSNSILEEATKRDDHEEEEDGLMLLSL 141
Lambda K H
0.306 0.124 0.330
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171470346
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40