bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2227_orf1
Length=121
Score E
Sequences producing significant alignments: (Bits) Value
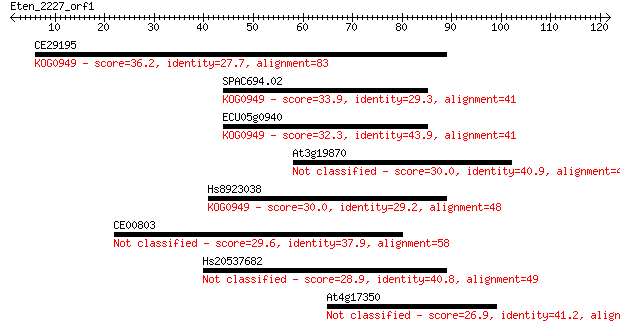
CE29195 36.2 0.017
SPAC694.02 33.9 0.079
ECU05g0940 32.3 0.25
At3g19870 30.0 0.97
Hs8923038 30.0 1.2
CE00803 29.6 1.6
Hs20537682 28.9 2.7
At4g17350 26.9 8.4
> CE29195
Length=1714
Score = 36.2 bits (82), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 23/86 (26%), Positives = 34/86 (39%), Gaps = 10/86 (11%)
Query 6 YFYLPLLSSSSSNH---PAPSWPNKDSKNASSRRTTELLAANSYLSDLYVHGKLARVASE 62
+ PL S + P + KD + +R N++ D YVHG +
Sbjct 1601 FLVAPLFDQYSHDESFLPVIDFNKKDHRGRKIQR-------NAFAYDFYVHGSRNMLMDV 1653
Query 63 NGVPPEQLWLLLQHFVLSLSRLSKGL 88
NG+ W LL F L RL+ G+
Sbjct 1654 NGLHVSVAWFLLHDFAAILERLAIGV 1679
> SPAC694.02
Length=1717
Score = 33.9 bits (76), Expect = 0.079, Method: Compositional matrix adjust.
Identities = 12/41 (29%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 44 NSYLSDLYVHGKLARVASENGVPPEQLWLLLQHFVLSLSRL 84
N+YL D + HG + + +N + ++W +L F L L+ +
Sbjct 1587 NAYLLDFFTHGSVDMLIEQNNLKQSEIWFVLNDFSLVLATI 1627
> ECU05g0940
Length=1337
Score = 32.3 bits (72), Expect = 0.25, Method: Composition-based stats.
Identities = 18/44 (40%), Positives = 27/44 (61%), Gaps = 3/44 (6%)
Query 44 NSYLSDLYVHGKLARVASENGVPPEQLW---LLLQHFVLSLSRL 84
NSYL D + HG +R+A+EN V +L+ ++ VLSL +L
Sbjct 1264 NSYLLDFFRHGSWSRIAAENRVGTGELFKSLSVVNSVVLSLIKL 1307
> At3g19870
Length=1117
Score = 30.0 bits (66), Expect = 0.97, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 24/47 (51%), Gaps = 3/47 (6%)
Query 58 RVASENGVPPEQLWLLLQHFVLSLSR---LSKGLLLLQQQQQQQGGC 101
R+A + +PP +L LL FV+SL L GL L Q + G C
Sbjct 540 RIAENDTIPPSRLIELLTKFVISLVEKRGLDVGLKLWDQGTEVLGIC 586
> Hs8923038
Length=391
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 25/48 (52%), Gaps = 0/48 (0%)
Query 41 LAANSYLSDLYVHGKLARVASENGVPPEQLWLLLQHFVLSLSRLSKGL 88
++ N+Y D Y HG L + +N + + LL+ F L++ +S L
Sbjct 315 MSLNAYALDFYKHGSLIGLVQDNRMNEGDAYYLLKDFALTIKSISVSL 362
> CE00803
Length=222
Score = 29.6 bits (65), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 31/63 (49%), Gaps = 9/63 (14%)
Query 22 PSWPNKDSKNASSRRTTELLAANSYLSDLYVHGKLARVASENGVP-----PEQLWLLLQH 76
P W KD S R ++ L A L+ L+V G +A E+ VP P+++ LQH
Sbjct 60 PHWKTKD---FPSSRISQHLVATMNLNSLFVAGGFGDLAVEDAVPSATGDPKKV-SHLQH 115
Query 77 FVL 79
F L
Sbjct 116 FGL 118
> Hs20537682
Length=653
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 20/49 (40%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query 40 LLAANSYLSDLYVHGKLARVASENGVPPEQLWLLLQHFVLSLSRLSKGL 88
LLA N + S+ V KL R AS +GVPP + L Q L S++ L
Sbjct 602 LLAGN-HSSEPKVSCKLGRSASTSGVPPPSVTPLRQSSDLQQSQVPSSL 649
> At4g17350
Length=383
Score = 26.9 bits (58), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 65 VPPEQLWLLLQHFVLSLSRLSKGLLLLQQQQQQQ 98
P E + L + + LS S +SK L L +QQQ Q
Sbjct 24 TPKEPMEFLSRSWSLSTSEISKALALKHRQQQDQ 57
Lambda K H
0.317 0.130 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1198419392
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40