bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2233_orf3
Length=263
Score E
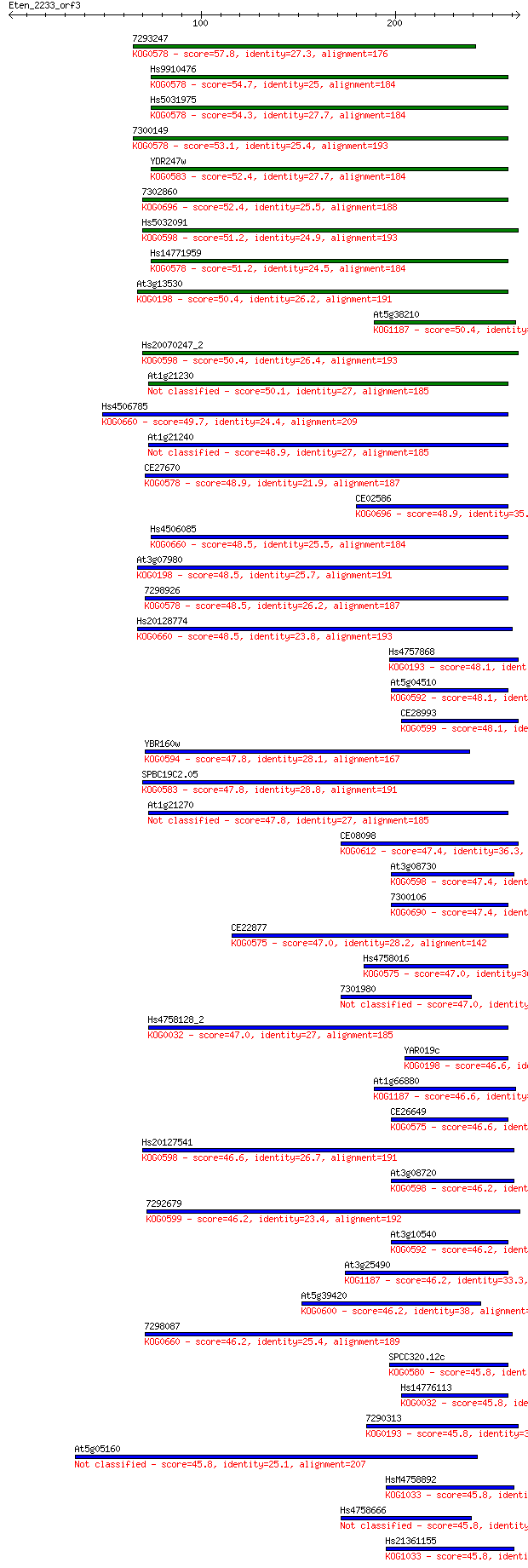
Sequences producing significant alignments: (Bits) Value
7293247 57.8 2e-08
Hs9910476 54.7 2e-07
Hs5031975 54.3 3e-07
7300149 53.1 6e-07
YDR247w 52.4 8e-07
7302860 52.4 1e-06
Hs5032091 51.2 2e-06
Hs14771959 51.2 2e-06
At3g13530 50.4 4e-06
At5g38210 50.4 4e-06
Hs20070247_2 50.4 4e-06
At1g21230 50.1 4e-06
Hs4506785 49.7 6e-06
At1g21240 48.9 1e-05
CE27670 48.9 1e-05
CE02586 48.9 1e-05
Hs4506085 48.5 1e-05
At3g07980 48.5 1e-05
7298926 48.5 1e-05
Hs20128774 48.5 1e-05
Hs4757868 48.1 2e-05
At5g04510 48.1 2e-05
CE28993 48.1 2e-05
YBR160w 47.8 2e-05
SPBC19C2.05 47.8 2e-05
At1g21270 47.8 3e-05
CE08098 47.4 3e-05
At3g08730 47.4 3e-05
7300106 47.4 3e-05
CE22877 47.0 4e-05
Hs4758016 47.0 4e-05
7301980 47.0 4e-05
Hs4758128_2 47.0 4e-05
YAR019c 46.6 5e-05
At1g66880 46.6 6e-05
CE26649 46.6 6e-05
Hs20127541 46.6 6e-05
At3g08720 46.2 6e-05
7292679 46.2 6e-05
At3g10540 46.2 6e-05
At3g25490 46.2 7e-05
At5g39420 46.2 7e-05
7298087 46.2 7e-05
SPCC320.12c 45.8 8e-05
Hs14776113 45.8 9e-05
7290313 45.8 9e-05
At5g05160 45.8 1e-04
HsM4758892 45.8 1e-04
Hs4758666 45.8 1e-04
Hs21361155 45.8 1e-04
> 7293247
Length=575
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 48/182 (26%), Positives = 74/182 (40%), Gaps = 40/182 (21%)
Query 65 RMRIMFLDVLGAGGIGVVLSAVDLSSKRRIAVKVCRLRSRSRRGPQYDQDVLKRRVQQEI 124
R + + +G G G V A D S+ R++AVK LR + RR +++ V+ R
Sbjct 365 RENLDHFNKIGEGSTGTVCIATDKSTGRQVAVKKMDLRKQQRRELLFNEVVIMR------ 418
Query 125 SLWKYVPPALDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPGDLLS 184
D P I+ T + + + WVV + G L+
Sbjct 419 -------------------------DYHHP--NIVETYSSFLVNDELWVVMEYLEGGALT 451
Query 185 ISEVWKGRM------PVRIEVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCT 238
V RM V + K + YLH+ G++H DIK+ + + DGR+ L D G C
Sbjct 452 -DIVTHSRMDEEQIATVCKQCLKALAYLHSQGVIHRDIKSDSILLAADGRVKLSDFGFCA 510
Query 239 PA 240
P
Sbjct 511 PG 512
> Hs9910476
Length=681
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 46/192 (23%), Positives = 85/192 (44%), Gaps = 41/192 (21%)
Query 74 LGAGGIGVVLSAVDLSSKRRIAVKVCRLRSRSRRGPQYDQDVLKRRVQQEISLWKYVPPA 133
+G G G+V A + S R++AVK+ LR + RR +++ V+ R Q
Sbjct 413 IGEGSTGIVCLAREKHSGRQVAVKMMDLRKQQRRELLFNEVVIMRDYQH----------- 461
Query 134 LDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPG----DLLSISEVW 189
+++E + + + + WV+ + G D++S +
Sbjct 462 ---------------FNVVEMYKSYL-------VGEELWVLMEFLQGGALTDIVSQVRLN 499
Query 190 KGRMPVRIEVAKQVM-YLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTPANQNT---E 245
+ ++ E Q + YLHA G++H DIK+ + + DGR+ L D G C +++ +
Sbjct 500 EEQIATVCEAVLQALAYLHAQGVIHRDIKSDSILLTLDGRVKLSDFGFCAQISKDVPKRK 559
Query 246 CLNGTRRYLPPE 257
L GT ++ PE
Sbjct 560 SLVGTPYWMAPE 571
> Hs5031975
Length=591
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 51/187 (27%), Positives = 80/187 (42%), Gaps = 31/187 (16%)
Query 74 LGAGGIGVVLSAVDLSSKRRIAVKVCRLRSRSRRGPQYDQDVLKRRVQQEISLWKYVPPA 133
+G G G+V A SS + +AVK LR + RR +++ V+ R Q E + Y
Sbjct 327 IGEGSTGIVCIATVRSSGKLVAVKKMDLRKQQRRELLFNEVVIMRDYQHENVVEMYNSYL 386
Query 134 LDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPGDLLSISEVWKGRM 193
+ + W VVM E +E TD + E + V
Sbjct 387 VGDELW-----VVM-----EFLEGGALTDIVTHTRMNEEQIAAVC--------------- 421
Query 194 PVRIEVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTPANQNT---ECLNGT 250
+ V + + LHA G++H DIK+ + + HDGR+ L D G C ++ + L GT
Sbjct 422 ---LAVLQALSVLHAQGVIHRDIKSDSILLTHDGRVKLSDFGFCAQVSKEVPRRKSLVGT 478
Query 251 RRYLPPE 257
++ PE
Sbjct 479 PYWMAPE 485
> 7300149
Length=569
Score = 53.1 bits (126), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 49/200 (24%), Positives = 87/200 (43%), Gaps = 38/200 (19%)
Query 65 RMRIMFLDVLGAGGIGVVLSAVDLSSKRRIAVKVCRLRSRSRRGPQYDQDVLKRRVQQEI 124
R R +G G G+V A DL ++ ++AVK ++++S + D + + RV ++
Sbjct 290 RERYKTTQEVGKGASGIVFIAADLQNESQVAVKTIDMKNQSSK----DLILTEIRVLKDF 345
Query 125 SLWKYVPPALDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPGDLLS 184
+ K + LD L+EP +++ WVV + G L+
Sbjct 346 N-HKNLVNFLDAY-------------LLEPEDQL-------------WVVMEYMDGGPLT 378
Query 185 ---ISEVWKGRMPVRI--EVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCT- 238
V K R + E + +LHA G++H DIK+ N + DG + + D G C
Sbjct 379 DVVTETVMKERQIACVCRETLYAISFLHAKGIIHRDIKSDNVLLGMDGSVKVTDFGFCAN 438
Query 239 -PANQNTECLNGTRRYLPPE 257
++ + + GT ++ PE
Sbjct 439 IEGDEKRQTMVGTPYWMAPE 458
> YDR247w
Length=461
Score = 52.4 bits (124), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 51/214 (23%), Positives = 97/214 (45%), Gaps = 33/214 (15%)
Query 74 LGAGGIGVVLSAVDLSSKRRIAVK-VCRLRSRSRRGPQYDQDVLKRRVQQEISLWKYVP- 131
+G G G+V A+D+ + R+ A+K V + S+ + + K V+ + L K
Sbjct 18 IGEGAYGLVYRALDIRTDRQYAIKAVVQSYGVSKEADMGNDKIHKNSVKLQKKLAKLFKE 77
Query 132 -------PALDVQQWSQISQ-----------VVMPLDLIEPIEKI-IHTDTESSIYSQEW 172
P++D++ +S+ + + L + + IH +S++ + +
Sbjct 78 SKNVVRVPSIDLESIENMSEEDFKKLPHYKEISLHLRVHHHKNIVTIHEVLQSAVCT--F 135
Query 173 VVFDVFPGDLL-SISE----VWKGRM--PVRIEVAKQVMYLHAMGLVHSDIKAPNFFVHH 225
+V D +P DL SI + V G + V +++ + Y H G+ H DIK N +
Sbjct 136 IVMDYYPTDLFTSIVDNRHFVTNGLLVKKVFLQICSALNYCHEHGIYHCDIKPENLLLDT 195
Query 226 DGRIYLGDNGLCTPAN--QNTECLNGTRRYLPPE 257
+ ++L D GL T + + C+ G+ Y+PPE
Sbjct 196 EDNVFLCDFGLSTTSTYIKPNVCI-GSSYYMPPE 228
> 7302860
Length=679
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 48/196 (24%), Positives = 82/196 (41%), Gaps = 36/196 (18%)
Query 70 FLDVLGAGGIGVVLSAVDLSSKRRIAVKVCRLRSRSRRGPQYDQDVLKRRVQQEISLWKY 129
F+ VLG G G VL A S+ A+K+ + + D DV +++ +
Sbjct 352 FIKVLGKGSFGKVLLAERKGSEELYAIKILKKDVIIQ-----DDDVECTMIEKRVLALGE 406
Query 130 VPPALDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPGDLL-SISEV 188
PP L Q+ +D + + E+V GDL+ I +
Sbjct 407 KPPFL-----VQLHSCFQTMDRL--------------FFVMEYVN----GGDLMFQIQQF 443
Query 189 WKGRMPVRI----EVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTP---AN 241
K + PV + E+A + +LH G+++ D+K N + DG + + D G+C +
Sbjct 444 GKFKEPVAVFYAAEIAAGLFFLHTKGILYRDLKLDNVLLDADGHVKIADFGMCKENIVGD 503
Query 242 QNTECLNGTRRYLPPE 257
+ T+ GT Y+ PE
Sbjct 504 KTTKTFCGTPDYIAPE 519
> Hs5032091
Length=431
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 48/200 (24%), Positives = 75/200 (37%), Gaps = 34/200 (17%)
Query 70 FLDVLGAGGIGVVLSAVDLSSKRRIAVKVCR----LRSRSRRGPQYDQDVLKRRVQQEIS 125
FL V+G G G VL A + + AVKV + L+ + + +++VL + V+
Sbjct 100 FLKVIGKGSFGKVLLARHKAEEVFYAVKVLQKKAILKKKEEKHIMSERNVLLKNVKHPFL 159
Query 126 LWKYVPPALDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPGDLLSI 185
+ + + ++ LD I E H E +
Sbjct 160 VGLHF-------SFQTADKLYFVLDYINGGELFYHLQRERCFLEPRARFYAA-------- 204
Query 186 SEVWKGRMPVRIEVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTPA---NQ 242
E+A + YLH++ +V+ D+K N + G I L D GLC N
Sbjct 205 ------------EIASALGYLHSLNIVYRDLKPENILLDSQGHIVLTDFGLCKENIEHNS 252
Query 243 NTECLNGTRRYLPPENMRCQ 262
T GT YL PE + Q
Sbjct 253 TTSTFCGTPEYLAPEVLHKQ 272
> Hs14771959
Length=719
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 45/193 (23%), Positives = 78/193 (40%), Gaps = 43/193 (22%)
Query 74 LGAGGIGVVLSAVDLSSKRRIAVKVCRLRSRSRRGPQYDQDVLKRRVQQEISLWKYVPPA 133
+G G G+V A + + +++AVK LR + RR +++ V+ R +
Sbjct 455 IGEGSTGIVCIATEKHTGKQVAVKKMDLRKQQRRELLFNEVVIMRDYHHD---------- 504
Query 134 LDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPGDLLSISEVWKGRM 193
++ + + + WVV + G L+ V RM
Sbjct 505 -----------------------NVVDMYSSYLVGDELWVVMEFLEGGALT-DIVTHTRM 540
Query 194 ------PVRIEVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTPANQNT--- 244
V + V + + YLH G++H DIK+ + + DGRI L D G C ++
Sbjct 541 NEEQIATVCLSVLRALSYLHNQGVIHRDIKSDSILLTSDGRIKLSDFGFCAQVSKEVPKR 600
Query 245 ECLNGTRRYLPPE 257
+ L GT ++ PE
Sbjct 601 KSLVGTPYWMAPE 613
> At3g13530
Length=1368
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 50/196 (25%), Positives = 89/196 (45%), Gaps = 30/196 (15%)
Query 67 RIMFLDVLGAGGIGVVLSAVDLSSKRRIAVKVCRLRSRSRRGPQYDQDVLKRRVQQEISL 126
+ M D +G G G V +DL + +A+K L + + +D+ + QEI L
Sbjct 19 KYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIVQ------EDL--NTIMQEIDL 70
Query 127 WKYVPPALDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPGDLLS-- 184
K + ++ ++ S+ L +I +E + + + I + F FP L++
Sbjct 71 LKNLNHK-NIVKYLGSSKTKTHLHII--LEYVENGSLANIIKPNK---FGPFPESLVAVY 124
Query 185 ISEVWKGRMPVRIEVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTPANQ-- 242
I++V +G ++YLH G++H DIK N +G + L D G+ T N+
Sbjct 125 IAQVLEG-----------LVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLNEAD 173
Query 243 -NTECLNGTRRYLPPE 257
NT + GT ++ PE
Sbjct 174 VNTHSVVGTPYWMAPE 189
> At5g38210
Length=960
Score = 50.4 bits (119), Expect = 4e-06, Method: Composition-based stats.
Identities = 25/77 (32%), Positives = 40/77 (51%), Gaps = 4/77 (5%)
Query 189 WKGRMPVRIEVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCT--PANQN--T 244
W R+ + IE A + YLHA G++H D+K N + + ++ + D GL P +Q +
Sbjct 730 WPARLQIAIETASALSYLHASGIIHRDVKTTNILLDSNYQVKVADFGLSRLFPMDQTHIS 789
Query 245 ECLNGTRRYLPPENMRC 261
GT Y+ PE +C
Sbjct 790 TAPQGTPGYVDPEYYQC 806
> Hs20070247_2
Length=378
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 51/200 (25%), Positives = 79/200 (39%), Gaps = 34/200 (17%)
Query 70 FLDVLGAGGIGVVLSAVDLSSKRRIAVKVCR----LRSRSRRGPQYDQDVLKRRVQQEIS 125
FL V+G G G VL A + AVKV + L + ++ +++VL + V+
Sbjct 46 FLKVIGKGSFGKVLLAKRKLDGKVYAVKVLQKKIVLNRKEQKHIMAERNVLLKNVKHPFL 105
Query 126 LWKYVPPALDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPGDLLSI 185
+ + + ++ LD + E H E S FP
Sbjct 106 VGLHY-------SFQTTEKLYFVLDFVNGGELFFHLQRERS-----------FPEH---- 143
Query 186 SEVWKGRMPVRIEVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTPA---NQ 242
+ R E+A + YLH++ +V+ D+K N V G + L D GLC +
Sbjct 144 ----RARF-YAAEIASALGYLHSIKIVYRDLKPENILVDSVGHVVLTDFGLCKEGIAISD 198
Query 243 NTECLNGTRRYLPPENMRCQ 262
T GT YL PE +R Q
Sbjct 199 TTTTFCGTPEYLAPEVIRKQ 218
> At1g21230
Length=733
Score = 50.1 bits (118), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 50/192 (26%), Positives = 79/192 (41%), Gaps = 32/192 (16%)
Query 73 VLGAGGIGVVLSAVDLSSKRRIAVKVCRLRSRSRRGPQYDQDVLKRRVQQEISLWKYVPP 132
+LG GG G V + L +A+K RL RS+ Q+ +VL ++ K +
Sbjct 413 ILGQGGQGTVYKGI-LQDNSIVAIKKARLGDRSQV-EQFINEVLVLSQINHRNVVKLLGC 470
Query 133 ALDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPGDLLSISEVWKGR 192
L+ + +PL + E I +FD G + S W+ R
Sbjct 471 CLETE---------VPLLVYEFISS--------------GTLFDHLHGSMFDSSLTWEHR 507
Query 193 MPVRIEVAKQVMYLHAMG---LVHSDIKAPNFFVHHDGRIYLGDNGLC--TPANQN--TE 245
+ + IEVA + YLH+ ++H D+K N + + + D G P +Q T
Sbjct 508 LRIAIEVAGTLAYLHSYASIPIIHRDVKTANILLDENLTAKVADFGASRLIPMDQEQLTT 567
Query 246 CLNGTRRYLPPE 257
+ GT YL PE
Sbjct 568 MVQGTLGYLDPE 579
> Hs4506785
Length=367
Score = 49.7 bits (117), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 51/213 (23%), Positives = 98/213 (46%), Gaps = 31/213 (14%)
Query 49 GFNKEVVVTSAARRGGRMRIMFLDV--LGAGGIGVVLSAVDLSSKRRIAVKVCRLRSRSR 106
GF ++ V +A +R ++ D+ +G+G G V SAVD + ++A+K +
Sbjct 10 GFYRQEVTKTA----WEVRAVYRDLQPVGSGAYGAVCSAVDGRTGAKVAIK--------K 57
Query 107 RGPQYDQDVLKRRVQQEISLWKYVPPALDVQQWSQISQVVMPLDLIEPIEKIIHTDTESS 166
+ ++ +R +E+ L K++ + V+ LD+ P E + D +
Sbjct 58 LYRPFQSELFAKRAYRELRLLKHM----------RHENVIGLLDVFTPDETL---DDFTD 104
Query 167 IY-SQEWVVFDVFPGDLLSISEVWKGRMPVRI-EVAKQVMYLHAMGLVHSDIKAPNFFVH 224
Y ++ D+ G L+ ++ + R+ + ++ K + Y+HA G++H D+K N V+
Sbjct 105 FYLVMPFMGTDL--GKLMKHEKLGEDRIQFLVYQMLKGLRYIHAAGIIHRDLKPGNLAVN 162
Query 225 HDGRIYLGDNGLCTPANQNTECLNGTRRYLPPE 257
D + + D GL A+ TR Y PE
Sbjct 163 EDCELKILDFGLARQADSEMTGYVVTRWYRAPE 195
> At1g21240
Length=741
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 50/192 (26%), Positives = 78/192 (40%), Gaps = 32/192 (16%)
Query 73 VLGAGGIGVVLSAVDLSSKRRIAVKVCRLRSRSRRGPQYDQDVLKRRVQQEISLWKYVPP 132
+LG GG G V + L +A+K RL + SR+ Q+ +VL ++ K +
Sbjct 420 ILGQGGQGTVYKGI-LPDNTIVAIKKARL-ADSRQVDQFIHEVLVLSQINHRNVVKILGC 477
Query 133 ALDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPGDLLSISEVWKGR 192
L+ + +PL + E I +FD G + S W+ R
Sbjct 478 CLETE---------VPLLVYEFITN--------------GTLFDHLHGSIFDSSLTWEHR 514
Query 193 MPVRIEVAKQVMYLHAMG---LVHSDIKAPNFFVHHDGRIYLGDNGLC----TPANQNTE 245
+ + IEVA + YLH+ ++H DIK N + + + D G Q T
Sbjct 515 LRIAIEVAGTLAYLHSSASIPIIHRDIKTANILLDENLTAKVADFGASKLIPMDKEQLTT 574
Query 246 CLNGTRRYLPPE 257
+ GT YL PE
Sbjct 575 MVQGTLGYLDPE 586
> CE27670
Length=572
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 41/195 (21%), Positives = 86/195 (44%), Gaps = 41/195 (21%)
Query 71 LDVLGAGGIGVVLSAVDLSSKRRIAVKVCRLRSRSRRGPQYDQDVLKRRVQQEISLWKYV 130
+D +G+G G V +A+++S++ +A+K L+ + ++ ++ +L R + ++ Y+
Sbjct 298 VDKIGSGASGSVYTAIEISTEAEVAIKQMNLKDQPKKELIINE-ILVMRENKHANIVNYL 356
Query 131 PPALDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPG----DLLSIS 186
L + + WVV + G D+++
Sbjct 357 DSYL--------------------------------VCDELWVVMEYLAGGSLTDVVTEC 384
Query 187 EVWKGRMP-VRIEVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLC---TPANQ 242
++ G + V EV + + +LH+ ++H DIK+ N + DG + L D G C +P +
Sbjct 385 QMEDGIIAAVCREVLQALEFLHSRHVIHRDIKSDNILLGMDGSVKLTDFGFCAQLSPEQR 444
Query 243 NTECLNGTRRYLPPE 257
+ GT ++ PE
Sbjct 445 KRTTMVGTPYWMAPE 459
> CE02586
Length=861
Score = 48.9 bits (115), Expect = 1e-05, Method: Composition-based stats.
Identities = 28/86 (32%), Positives = 46/86 (53%), Gaps = 8/86 (9%)
Query 180 GDLL-SISEVWKGRMPVRI----EVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDN 234
GDL+ I +V K + PV + E+A + +LH+ G+++ D+K N + DG I + D
Sbjct 497 GDLMYQIQQVGKFKEPVAVFYAAEIAVGLFFLHSKGIIYRDLKLDNVMLERDGHIKITDF 556
Query 235 GLCTP---ANQNTECLNGTRRYLPPE 257
G+C + T+ GT Y+ PE
Sbjct 557 GMCKENIFGDATTKTFCGTPDYIAPE 582
> Hs4506085
Length=365
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 47/188 (25%), Positives = 83/188 (44%), Gaps = 30/188 (15%)
Query 74 LGAGGIGVVLSAVDLSSKRRIAVKVCR--LRSRSRRGPQYDQDVLKRRVQQE--ISLWKY 129
+G+G G V SA+D S ++A+K +S Y + +L + +Q E I L
Sbjct 31 VGSGAYGSVCSAIDKRSGEKVAIKKLSRPFQSEIFAKRAYRELLLLKHMQHENVIGLLDV 90
Query 130 VPPALDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPGDLLSISEVW 189
PA ++ + +VMP + ++KI+ + +S+E + + V+
Sbjct 91 FTPASSLRNFYDFY-LVMPF-MQTDLQKIMGME-----FSEEKIQYLVY----------- 132
Query 190 KGRMPVRIEVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTPANQNTECLNG 249
++ K + Y+H+ G+VH D+K N V+ D + + D GL A+
Sbjct 133 --------QMLKGLKYIHSAGVVHRDLKPGNLAVNEDCELKILDFGLARHADAEMTGYVV 184
Query 250 TRRYLPPE 257
TR Y PE
Sbjct 185 TRWYRAPE 192
> At3g07980
Length=1367
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 49/196 (25%), Positives = 85/196 (43%), Gaps = 30/196 (15%)
Query 67 RIMFLDVLGAGGIGVVLSAVDLSSKRRIAVKVCRLRSRSRRGPQYDQDVLKRRVQQEISL 126
+ M D +G G G V +DL + +A+K L + + +D+ + QEI L
Sbjct 19 KYMLGDEIGKGAYGRVYIGLDLENGDFVAIKQVSLENIGQ------EDL--NTIMQEIDL 70
Query 127 WKYVPPALDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPGDLLS-- 184
K + V+ + ++E +E + ++I F FP L++
Sbjct 71 LKNLNHKNIVKYLGSLKTKTHLHIILEYVE----NGSLANIIKPNK--FGPFPESLVTVY 124
Query 185 ISEVWKGRMPVRIEVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTPANQ-- 242
I++V +G ++YLH G++H DIK N +G + L D G+ T N+
Sbjct 125 IAQVLEG-----------LVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLNEAD 173
Query 243 -NTECLNGTRRYLPPE 257
NT + GT ++ PE
Sbjct 174 FNTHSVVGTPYWMAPE 189
> 7298926
Length=704
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 49/196 (25%), Positives = 87/196 (44%), Gaps = 43/196 (21%)
Query 71 LDVLGAGGIGVVLSAVDLSSKRRIAVKVCRLRSRSRRGPQYDQDVLKRRVQQEISLWKYV 130
++ +G G G V +A++ S+ +A+K L + + K + EI
Sbjct 433 MEKIGQGASGTVYTAIESSTGMEVAIKQMNLSQQPK----------KELIINEIL----- 477
Query 131 PPALDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQE-WVVFDVFPG----DLLSI 185
V + ++ VV LD S + S+E WVV + PG D+++
Sbjct 478 -----VMRENKHPNVVNYLD--------------SYLVSEELWVVMEYLPGGSLTDVVTE 518
Query 186 SEVWKGRMP-VRIEVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLC---TPAN 241
+ + +G++ V EV + + +LHA ++H DIK+ N + DG + L D G C +P
Sbjct 519 TCMDEGQIAAVCREVLQALEFLHANQVIHRDIKSDNILLGLDGSVKLTDFGFCAQISPEQ 578
Query 242 QNTECLNGTRRYLPPE 257
+ GT ++ PE
Sbjct 579 SKRTTMVGTPYWMAPE 594
> Hs20128774
Length=364
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 46/197 (23%), Positives = 86/197 (43%), Gaps = 29/197 (14%)
Query 67 RIMFLDVLGAGGIGVVLSAVDLSSKRRIAVKVCRLRSRSRRGPQYDQDVLKRRVQQEISL 126
R+ L +G+G G V SA D ++++AVK + + + RR +E+ L
Sbjct 23 RLQGLRPVGSGAYGSVCSAYDARLRQKVAVK--------KLSRPFQSLIHARRTYRELRL 74
Query 127 WKYVPPALDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPGDLLSIS 186
K++ + V+ LD+ P I +S+ ++V + DL +I
Sbjct 75 LKHL----------KHENVIGLLDVFTPATSI-------EDFSEVYLVTTLMGADLNNIV 117
Query 187 EVW----KGRMPVRIEVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTPANQ 242
+ + + ++ + + Y+H+ G++H D+K N V+ D + + D GL A++
Sbjct 118 KCQALSDEHVQFLVYQLLRGLKYIHSAGIIHRDLKPSNVAVNEDCELRILDFGLARQADE 177
Query 243 NTECLNGTRRYLPPENM 259
TR Y PE M
Sbjct 178 EMTGYVATRWYRAPEIM 194
> Hs4757868
Length=765
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 26/75 (34%), Positives = 42/75 (56%), Gaps = 9/75 (12%)
Query 197 IEVAKQVM----YLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCT-----PANQNTECL 247
I++A+Q YLHA ++H D+K+ N F+H D + +GD GL T + E L
Sbjct 553 IDIARQTAQGMDYLHAKSIIHRDLKSNNIFLHEDLTVKIGDFGLATVKSRWSGSHQFEQL 612
Query 248 NGTRRYLPPENMRCQ 262
+G+ ++ PE +R Q
Sbjct 613 SGSILWMAPEVIRMQ 627
> At5g04510
Length=488
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 35/73 (47%), Gaps = 13/73 (17%)
Query 198 EVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTP------------ANQNTE 245
EV + Y+H+MGL+H DIK N + DG I + D G P A+ +
Sbjct 147 EVVDALEYIHSMGLIHRDIKPENLLLTSDGHIKIADFGSVKPMQDSQITVLPNAASDDKA 206
Query 246 C-LNGTRRYLPPE 257
C GT Y+PPE
Sbjct 207 CTFVGTAAYVPPE 219
> CE28993
Length=370
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 32/62 (51%), Gaps = 2/62 (3%)
Query 203 VMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCT--PANQNTECLNGTRRYLPPENMR 260
V Y+HA +VH D+K N + RI + D G T P + L GT YL PE +R
Sbjct 33 VEYMHARDIVHRDLKLENILCIDEERIVISDFGFATRIPRGKKLRDLCGTPGYLAPETIR 92
Query 261 CQ 262
CQ
Sbjct 93 CQ 94
> YBR160w
Length=298
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 47/178 (26%), Positives = 73/178 (41%), Gaps = 41/178 (23%)
Query 71 LDVLGAGGIGVVLSAVDL---SSKRRIAVKVCRLRSRSRRGPQYDQDVLKRRVQQEISLW 127
L+ +G G GVV A+DL +R +A+K RL S P +EISL
Sbjct 11 LEKVGEGTYGVVYKALDLRPGQGQRVVALKKIRLESEDEGVPS--------TAIREISLL 62
Query 128 KYVPPALDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPGDLLSISE 187
K + D I + I+H+D + ++VF+ DL E
Sbjct 63 KELKD-----------------DNIVRLYDIVHSDAH-----KLYLVFEFLDLDLKRYME 100
Query 188 VWKGRMPVRIEVAKQVM--------YLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLC 237
P+ ++ K+ M Y H+ ++H D+K N ++ DG + LGD GL
Sbjct 101 GIPKDQPLGADIVKKFMMQLCKGIAYCHSHRILHRDLKPQNLLINKDGNLKLGDFGLA 158
> SPBC19C2.05
Length=470
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 55/200 (27%), Positives = 91/200 (45%), Gaps = 39/200 (19%)
Query 70 FLDVLGAGGIGVVLSAVDLSSKRRIAVKVCRLRSRSRRGPQYDQDVLKRRVQQEISLWKY 129
F+ ++GAG GVV A D+ AVK + + G Q L+ R E++L
Sbjct 20 FVSIIGAGAYGVVYKAEDIYDGTLYAVK-----ALCKDGLNEKQKKLQAR---ELALHAR 71
Query 130 VPPALDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFP-GDLLS-ISE 187
V S ++ + +++ +TE +IY VV P GDL + I+E
Sbjct 72 V---------SSHPYIIT-------LHRVL--ETEDAIY----VVLQYCPNGDLFTYITE 109
Query 188 --VWKGRM----PVRIEVAKQVMYLHAMGLVHSDIKAPNFFVHHD-GRIYLGDNGLCTPA 240
V++G V +++ V + H++G+ H D+K N V +D +YL D GL T
Sbjct 110 KKVYQGNSHLIKTVFLQLISAVEHCHSVGIYHRDLKPENIMVGNDVNTVYLADFGLATTE 169
Query 241 NQNTECLNGTRRYLPPENMR 260
+++ G+ Y+ PE R
Sbjct 170 PYSSDFGCGSLFYMSPECQR 189
> At1g21270
Length=732
Score = 47.8 bits (112), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 50/192 (26%), Positives = 77/192 (40%), Gaps = 32/192 (16%)
Query 73 VLGAGGIGVVLSAVDLSSKRRIAVKVCRLRSRSRRGPQYDQDVLKRRVQQEISLWKYVPP 132
+LG GG G V + L +A+K RL +RS+ Q+ +VL ++ K +
Sbjct 409 ILGQGGQGTVYKGI-LPDNSIVAIKKARLGNRSQV-EQFINEVLVLSQINHRNVVKVLGC 466
Query 133 ALDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPGDLLSISEVWKGR 192
L+ + +PL + E I +FD G L S W+ R
Sbjct 467 CLETE---------VPLLVYEFI--------------NSGTLFDHLHGSLYDSSLTWEHR 503
Query 193 MPVRIEVAKQVMYLHAMG---LVHSDIKAPNFFVHHDGRIYLGDNGLC----TPANQNTE 245
+ + EVA + YLH+ ++H DIK N + + + D G Q T
Sbjct 504 LRIATEVAGSLAYLHSSASIPIIHRDIKTANILLDKNLTAKVADFGASRLIPMDKEQLTT 563
Query 246 CLNGTRRYLPPE 257
+ GT YL PE
Sbjct 564 IVQGTLGYLDPE 575
> CE08098
Length=1173
Score = 47.4 bits (111), Expect = 3e-05, Method: Composition-based stats.
Identities = 33/102 (32%), Positives = 48/102 (47%), Gaps = 13/102 (12%)
Query 172 WVVFDVFPG-DLLS------ISEVWKGRMPVRIEVAKQVMYLHAMGLVHSDIKAPNFFVH 224
++V + PG DL++ +SE W E+ + + LH+MG +H D+K N +
Sbjct 142 YMVMEYMPGGDLVNLMTSYEVSEKWTRFYTA--EIVEALAALHSMGYIHRDVKPDNMLIS 199
Query 225 HDGRIYLGDNGLCTPANQN--TECLN--GTRRYLPPENMRCQ 262
G I L D G C N N C GT Y+ PE +R Q
Sbjct 200 ISGHIKLADFGTCVKMNANGVVRCSTAVGTPDYISPEVLRNQ 241
> At3g08730
Length=465
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 34/65 (52%), Gaps = 2/65 (3%)
Query 198 EVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTPANQNT--ECLNGTRRYLP 255
E+ V +LH G++H D+K N + DG + L D GL +NT + GT Y+
Sbjct 240 EIVSAVSHLHEKGIMHRDLKPENILMDTDGHVMLTDFGLAKEFEENTRSNSMCGTTEYMA 299
Query 256 PENMR 260
PE +R
Sbjct 300 PEIVR 304
> 7300106
Length=530
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 33/63 (52%), Gaps = 3/63 (4%)
Query 198 EVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLC---TPANQNTECLNGTRRYL 254
E+ + YLH+ G+++ D+K N + DG I + D GLC + T+ GT YL
Sbjct 291 EIISALGYLHSQGIIYRDLKLENLLLDKDGHIKVADFGLCKEDITYGRTTKTFCGTPEYL 350
Query 255 PPE 257
PE
Sbjct 351 APE 353
> CE22877
Length=632
Score = 47.0 bits (110), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 40/157 (25%), Positives = 76/157 (48%), Gaps = 21/157 (13%)
Query 116 LKRRVQQEISLWKYVPPALDVQQW-----SQISQVVMPLDLIEPIEKIIHTDTESSIYSQ 170
L+ + E+ K VP AL ++Q+ +Q Q+ L ++ + +S++Y
Sbjct 53 LRNKSTGELFAGKVVPKALLIKQYQRDKMAQEVQIHRNLQHRNVVKLYHFFEDKSNVY-- 110
Query 171 EWVVFDVFPGDLLSISEVWKGRMPVR--------IEVAKQVMYLHAMGLVHSDIKAPNFF 222
+ ++ P S+ E+ K R V ++ + V+YLH + +VH D+K N F
Sbjct 111 --ITLELCPRR--SLMELHKRRKAVTEPEARYFTYQIVEGVLYLHNLKIVHRDLKLGNLF 166
Query 223 VHHDGRIYLGDNGLCTPANQN--TECLNGTRRYLPPE 257
++ + ++ +GD GL T + + + L GT Y+ PE
Sbjct 167 LNDELQVKIGDFGLATTCDNDERKKTLCGTPNYIAPE 203
> Hs4758016
Length=607
Score = 47.0 bits (110), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 27/85 (31%), Positives = 43/85 (50%), Gaps = 11/85 (12%)
Query 184 SISEVWKGRMP-----VRI---EVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNG 235
S++ +WK R VR ++ + YLH G++H D+K NFF+ + + +GD G
Sbjct 107 SLAHIWKARHTLLEPEVRYYLRQILSGLKYLHQRGILHRDLKLGNFFITENMELKVGDFG 166
Query 236 LCT---PANQNTECLNGTRRYLPPE 257
L P Q + + GT Y+ PE
Sbjct 167 LAARLEPPEQRKKTICGTPNYVAPE 191
> 7301980
Length=1105
Score = 47.0 bits (110), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 30/74 (40%), Positives = 36/74 (48%), Gaps = 8/74 (10%)
Query 172 WVVFDVFPG-DLLS------ISEVWKGRMPVRIEVAKQVMYLHAMGLVHSDIKAPNFFVH 224
+ V D PG DL+S I E R + EV V +H MG +H DIK N +
Sbjct 794 YFVMDYIPGGDLMSLLIKLGIFEEELARFYI-AEVTCAVDSVHKMGFIHRDIKPDNILID 852
Query 225 HDGRIYLGDNGLCT 238
DG I L D GLCT
Sbjct 853 RDGHIKLTDFGLCT 866
> Hs4758128_2
Length=385
Score = 47.0 bits (110), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 50/190 (26%), Positives = 82/190 (43%), Gaps = 32/190 (16%)
Query 73 VLGAGGIGVVLSAVDLSSKRRIAVKVCRLRSRSRRGPQYDQDVLKRRVQQEISLWKYVP- 131
+G G VV V+ S+ R A+K+ ++ RG ++ +Q E+S+ + V
Sbjct 51 TIGDGNFAVVKECVERSTAREYALKI--IKKSKCRGKEH-------MIQNEVSILRRVKH 101
Query 132 PALDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPGDLLSISEVWKG 191
P + + I ++ +P +L +E + D +I S G L ++
Sbjct 102 PNIVLL----IEEMDVPTELYLVMELVKGGDLFDAITSTNKYTERDASGMLYNL------ 151
Query 192 RMPVRIEVAKQVMYLHAMGLVHSDIKAPNFFV--HHDG--RIYLGDNGLCTPANQNTECL 247
A + YLH++ +VH DIK N V H DG + LGD GL T + +
Sbjct 152 --------ASAIKYLHSLNIVHRDIKPENLLVYEHQDGSKSLKLGDFGLATIVDGPLYTV 203
Query 248 NGTRRYLPPE 257
GT Y+ PE
Sbjct 204 CGTPTYVAPE 213
> YAR019c
Length=974
Score = 46.6 bits (109), Expect = 5e-05, Method: Composition-based stats.
Identities = 21/53 (39%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 205 YLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTPANQNTECLNGTRRYLPPE 257
YLH G++H DIKA N + D + L D G+ T N + L GT ++ PE
Sbjct 136 YLHGEGVIHRDIKAANILLSADNTVKLADFGVSTIVNSSALTLAGTLNWMAPE 188
> At1g66880
Length=1286
Score = 46.6 bits (109), Expect = 6e-05, Method: Composition-based stats.
Identities = 24/77 (31%), Positives = 39/77 (50%), Gaps = 4/77 (5%)
Query 189 WKGRMPVRIEVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCT--PANQN--T 244
W R+ + IE A + +LH G++H DIK N + + ++ + D GL P +Q +
Sbjct 1055 WSTRLNIAIETASALSFLHIKGIIHRDIKTTNILLDDNYQVKVADFGLSRLFPMDQTHIS 1114
Query 245 ECLNGTRRYLPPENMRC 261
GT Y+ PE +C
Sbjct 1115 TAPQGTPGYVDPEYYQC 1131
> CE26649
Length=648
Score = 46.6 bits (109), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 36/62 (58%), Gaps = 2/62 (3%)
Query 198 EVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTPAN--QNTECLNGTRRYLP 255
++ V+YLH + ++H D+K N F++ D + +GD GL T N + + L GT Y+
Sbjct 144 QIVDGVLYLHDLNIIHRDMKLGNLFLNDDLVVKIGDFGLATTVNGDERKKTLCGTPNYIA 203
Query 256 PE 257
PE
Sbjct 204 PE 205
> Hs20127541
Length=427
Score = 46.6 bits (109), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 51/202 (25%), Positives = 74/202 (36%), Gaps = 42/202 (20%)
Query 70 FLDVLGAGGIGVVLSAVDLSSKRRIAVKVCRLRSRSRRGPQYDQDVLKRRVQQEISLWKY 129
FL V+G G G VL A S AVKV + +S +LK++ Q I +
Sbjct 97 FLKVIGKGNYGKVLLAKRKSDGAFYAVKVLQKKS-----------ILKKKEQSHIMAERS 145
Query 130 V------PPALDVQQWS--QISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPGD 181
V P L ++S ++ LD + E H E +
Sbjct 146 VLLKNVRHPFLVGLRYSFQTPEKLYFVLDYVNGGELFFHLQRERRFLEPRARFYAA---- 201
Query 182 LLSISEVWKGRMPVRIEVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTPA- 240
EVA + YLH++ +++ D+K N + G + L D GLC
Sbjct 202 ----------------EVASAIGYLHSLNIIYRDLKPENILLDCQGHVVLTDFGLCKEGV 245
Query 241 --NQNTECLNGTRRYLPPENMR 260
T GT YL PE +R
Sbjct 246 EPEDTTSTFCGTPEYLAPEVLR 267
> At3g08720
Length=471
Score = 46.2 bits (108), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 34/65 (52%), Gaps = 2/65 (3%)
Query 198 EVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTPANQNT--ECLNGTRRYLP 255
E+ V +LH G++H D+K N + DG + L D GL +NT + GT Y+
Sbjct 246 EIVSAVSHLHEKGIMHRDLKPENILMDVDGHVMLTDFGLAKEFEENTRSNSMCGTTEYMA 305
Query 256 PENMR 260
PE +R
Sbjct 306 PEIVR 310
> 7292679
Length=560
Score = 46.2 bits (108), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 45/197 (22%), Positives = 87/197 (44%), Gaps = 25/197 (12%)
Query 72 DVLGAGGIGVVLSAVDLSSKRRIAVKVCRLRSRSRRGPQYDQDVLKRRVQQEISLWKYV- 130
++LG G V ++ + + A K+ L + + G +L+ +QEIS+ + V
Sbjct 27 EILGRGISSTVRRCIEKETGKEFAAKIIDLGATTESGETNPYHMLEA-TRQEISILRQVM 85
Query 131 --PPALDVQQWSQISQVVMPLDLIEPIEKIIHTDTESSIYSQEWVVFDVFPGDLLSISEV 188
P +D+Q + V + + P ++ FD + ++++SE
Sbjct 86 GHPYIIDLQDVFESDAFVFLVFELCPKGEL----------------FD-YLTSVVTLSEK 128
Query 189 WKGRMPVRIEVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTPANQNTECLN 248
K R +R ++ + V Y+HA +VH D+K N + + + + D G + + N
Sbjct 129 -KTRTIMR-QIFEGVEYIHAKSIVHRDLKPENILLDENHNVKITDFGFAKQLQEGEKLTN 186
Query 249 --GTRRYLPPENMRCQL 263
GT YL PE ++C +
Sbjct 187 LCGTPGYLAPETLKCNM 203
> At3g10540
Length=483
Score = 46.2 bits (108), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 34/73 (46%), Gaps = 13/73 (17%)
Query 198 EVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTP------------ANQNTE 245
EV + Y+H MGL+H DIK N + DG I + D G P A+ +
Sbjct 148 EVVDALEYIHNMGLIHRDIKPENLLLTLDGHIKIADFGSVKPMQDSQITVLPNAASDDKA 207
Query 246 C-LNGTRRYLPPE 257
C GT Y+PPE
Sbjct 208 CTFVGTAAYVPPE 220
> At3g25490
Length=433
Score = 46.2 bits (108), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 28/91 (30%), Positives = 41/91 (45%), Gaps = 7/91 (7%)
Query 174 VFDVFPGDLLSISEVWKGRMPVRIEVAKQVMYLHAMG---LVHSDIKAPNFFVHHDGRIY 230
+FD G + S W+ R+ + IEVA + YLH+ ++H DIK N + +
Sbjct 189 LFDHLHGSMFVSSLTWEHRLEIAIEVAGAIAYLHSGASIPIIHRDIKTENILLDENLTAK 248
Query 231 LGDNGLC----TPANQNTECLNGTRRYLPPE 257
+ D G Q T + GT YL PE
Sbjct 249 VADFGASKLKPMDKEQLTTMVQGTLGYLDPE 279
> At5g39420
Length=576
Score = 46.2 bits (108), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 35/101 (34%), Positives = 51/101 (50%), Gaps = 13/101 (12%)
Query 152 IEPIEKIIHTDTESSIYSQEWVVFDVFPGDLLSISEVWKGRM--PVRIEVAKQVM----Y 205
I +E I+ + SSIY +VF+ DL +S R P KQ++ +
Sbjct 164 IMKLEGIVTSRASSSIY----LVFEYMEHDLAGLSSNPDIRFTEPQIKCYMKQLLWGLEH 219
Query 206 LHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLC---TPANQN 243
H G++H DIKA N V++ G + LGD GL TP+N+N
Sbjct 220 CHMRGVIHRDIKASNILVNNKGVLKLGDFGLANVVTPSNKN 260
> 7298087
Length=365
Score = 46.2 bits (108), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 48/194 (24%), Positives = 74/194 (38%), Gaps = 28/194 (14%)
Query 71 LDVLGAGGIGVVLSAVDLSSKRRIAVKVCR--LRSRSRRGPQYDQDVLKRRVQQE--ISL 126
L +G G G V AV + ++A+K +S Y + L + + E I L
Sbjct 27 LQPVGQGAYGQVCKAVVRGTSTKVAIKKLARPFQSAVHAKRTYRELRLLKHMDHENVIGL 86
Query 127 WKYVPPALDVQQWSQISQVVMPLDLIEP-IEKIIHTDTESSIYSQEWVVFDVFPGDLLSI 185
P Q QV M L++ + II T S + Q ++V+ + G
Sbjct 87 LDVFHPGQPADSLDQFQQVYMVTHLMDADLNNIIRTQKLSDDHVQ-FLVYQILRG----- 140
Query 186 SEVWKGRMPVRIEVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTPANQNTE 245
+ Y+H+ G++H D+K N V+ D + + D GL PA
Sbjct 141 -----------------LKYIHSAGVIHRDLKPSNIAVNEDCELRILDFGLARPAESEMT 183
Query 246 CLNGTRRYLPPENM 259
TR Y PE M
Sbjct 184 GYVATRWYRAPEIM 197
> SPCC320.12c
Length=384
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 26/63 (41%), Positives = 35/63 (55%), Gaps = 3/63 (4%)
Query 197 IEVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNG--LCTPANQNTECLNGTRRYL 254
++A + YLH ++H DIK N + DG I L D G + P+N+ T L GT YL
Sbjct 223 FQMANALSYLHKKHVIHRDIKPENILLGIDGEIKLSDFGWSVHAPSNRRT-TLCGTLDYL 281
Query 255 PPE 257
PPE
Sbjct 282 PPE 284
> Hs14776113
Length=424
Score = 45.8 bits (107), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 33/62 (53%), Gaps = 7/62 (11%)
Query 203 VMYLHAMGLVHSDIKAPNFFVHH---DGRIYLGDNGLCTPANQNTECL----NGTRRYLP 255
V YLHA+G+ H D+K N +H D +I + D GL + + +CL GT Y+
Sbjct 206 VRYLHALGITHRDLKPENLLYYHPGTDSKIIITDFGLASARKKGDDCLMKTTCGTPEYIA 265
Query 256 PE 257
PE
Sbjct 266 PE 267
> 7290313
Length=739
Score = 45.8 bits (107), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 31/91 (34%), Positives = 48/91 (52%), Gaps = 18/91 (19%)
Query 185 ISEVWKGRMPVRIEVAKQVM----YLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTP- 239
+SE K ++ I++ +QV YLHA ++H D+K+ N F+H D + +GD GL T
Sbjct 515 VSET-KFKLNTLIDIGRQVAQGMDYLHAKNIIHRDLKSNNIFLHEDLSVKIGDFGLATAK 573
Query 240 --------ANQNTECLNGTRRYLPPENMRCQ 262
ANQ T G+ ++ PE +R Q
Sbjct 574 TRWSGEKQANQPT----GSILWMAPEVIRMQ 600
> At5g05160
Length=640
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 52/210 (24%), Positives = 82/210 (39%), Gaps = 42/210 (20%)
Query 35 PELSKERALWRDAYGFNKEVVVTSAARRGGRMRIMFLDVLGAGGIGVVLSAVDLSSKRRI 94
PE +K R + F+ E ++ ++A +VLG G G AV +
Sbjct 325 PEKNKLFFFERCNHNFDLEDLLKASA-----------EVLGKGSFGTAYKAV---LEDTT 370
Query 95 AVKVCRLRSRSRRGPQYDQDVLKRRVQQEISLWKYVPPALDVQQWSQISQVVMPLDLIEP 154
AV V RLR +++Q Q EI V + +Q S V P
Sbjct 371 AVVVKRLREVVASKKEFEQ-------QMEI-----------VGKINQHSNFV-------P 405
Query 155 IEKIIHTDTESSI---YSQEWVVFDVFPGDLLSISEVWKGRMPVRIEVAKQVMYLHAMGL 211
+ ++ E + Y + +F + G+ W+ RM + +K + YLH++
Sbjct 406 LLAYYYSKDEKLLVYKYMTKGSLFGIMHGNRGDRGVDWETRMKIATGTSKAISYLHSLKF 465
Query 212 VHSDIKAPNFFVHHDGRIYLGDNGLCTPAN 241
VH DIK+ N + D L D L T N
Sbjct 466 VHGDIKSSNILLTEDLEPCLSDTSLVTLFN 495
> HsM4758892
Length=1115
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 40/81 (49%), Gaps = 15/81 (18%)
Query 195 VRIEVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTPANQNTECLN------ 248
+ +++A+ V +LH+ GL+H D+K N F D + +GD GL T +Q+ E
Sbjct 916 IFLQIAEAVEFLHSKGLMHRDLKPSNIFFTMDDVVKVGDFGLVTAMDQDEEEQTVLTPMP 975
Query 249 ---------GTRRYLPPENMR 260
GT+ Y+ PE +
Sbjct 976 AYARHTGQVGTKLYMSPEQIH 996
> Hs4758666
Length=1130
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 37/74 (50%), Gaps = 8/74 (10%)
Query 172 WVVFDVFPG-DLLSISEVWKGRMPVRI------EVAKQVMYLHAMGLVHSDIKAPNFFVH 224
+ V D PG D++S+ + G P + E+ V +H MG +H DIK N +
Sbjct 779 YFVMDYIPGGDMMSLL-IRMGIFPESLARFYIAELTCAVESVHKMGFIHRDIKPDNILID 837
Query 225 HDGRIYLGDNGLCT 238
DG I L D GLCT
Sbjct 838 RDGHIKLTDFGLCT 851
> Hs21361155
Length=1115
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 40/81 (49%), Gaps = 15/81 (18%)
Query 195 VRIEVAKQVMYLHAMGLVHSDIKAPNFFVHHDGRIYLGDNGLCTPANQNTECLN------ 248
+ +++A+ V +LH+ GL+H D+K N F D + +GD GL T +Q+ E
Sbjct 916 IFLQIAEAVEFLHSKGLMHRDLKPSNIFFTMDDVVKVGDFGLVTAMDQDEEEQTVLTPMP 975
Query 249 ---------GTRRYLPPENMR 260
GT+ Y+ PE +
Sbjct 976 AYARHTGQVGTKLYMSPEQIH 996
Lambda K H
0.322 0.138 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5578361662
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40