bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2235_orf2
Length=299
Score E
Sequences producing significant alignments: (Bits) Value
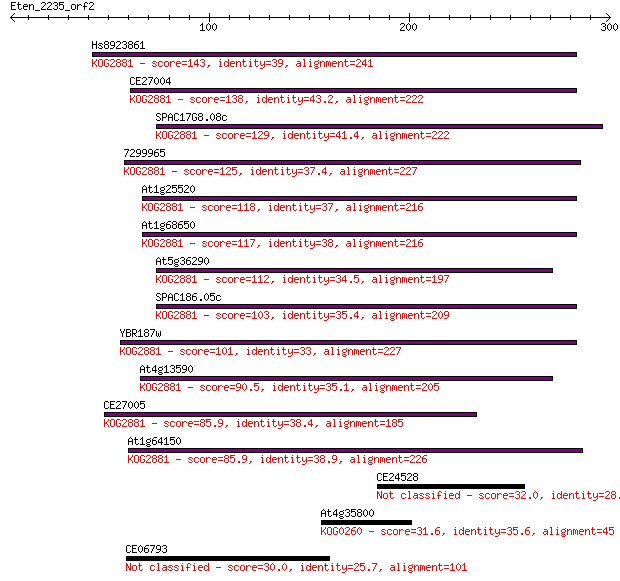
Hs8923861 143 4e-34
CE27004 138 2e-32
SPAC17G8.08c 129 7e-30
7299965 125 1e-28
At1g25520 118 1e-26
At1g68650 117 3e-26
At5g36290 112 8e-25
SPAC186.05c 103 3e-22
YBR187w 101 2e-21
At4g13590 90.5 4e-18
CE27005 85.9 9e-17
At1g64150 85.9 1e-16
CE24528 32.0 1.8
At4g35800 31.6 1.9
CE06793 30.0 6.1
> Hs8923861
Length=324
Score = 143 bits (361), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 94/263 (35%), Positives = 138/263 (52%), Gaps = 43/263 (16%)
Query 42 LFLKAVKASAGQVLPVPEGDISWSSAFVGTLFMIICSELGDKTFLITAVLSMRCTKVDSS 101
+F A + P + ++ + AFV + +II SELGDKTF I A+++MR
Sbjct 71 IFTPAAPVHTNKEDPATQTNLGFIHAFVAAISVIIVSELGDKTFFIAAIMAMRY------ 124
Query 102 KKSRPNEKRPGSRWHVFAGGFLALVVMTLMAAAGG---QVLPLLLKQELRTIFMVCLLVI 158
+R V AG LAL +MT ++ G V+P + + T+ L I
Sbjct 125 -----------NRLTVLAGAMLALGLMTCLSVLFGYATTVIPRVYTYYVSTV----LFAI 169
Query 159 FGAKMLHEA--AVYEENSDEPEELK-DLAQSEESMSQSRLF-------------VLARS- 201
FG +ML E +E +E EE++ +L + +E +++L V +
Sbjct 170 FGIRMLREGLKMSPDEGQEELEEVQAELKKKDEEFQRTKLLNGPGDVETGTSITVPQKKW 229
Query 202 --FFSPVFIQAASLTLMAEWGDRSQLATFALAADRSASAVCLGAILGHGLCTALAVIGGN 259
F SP+F+QA +LT +AEWGDRSQL T LAA V +G +GH LCT LAVIGG
Sbjct 230 LHFISPIFVQALTLTFLAEWGDRSQLTTIVLAAREDPYGVAVGGTVGHCLCTGLAVIGGR 289
Query 260 VLAKRISERVVLVLGGICFLSFA 282
++A++IS R V ++GGI FL+FA
Sbjct 290 MIAQKISVRTVTIIGGIVFLAFA 312
> CE27004
Length=297
Score = 138 bits (347), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 96/247 (38%), Positives = 129/247 (52%), Gaps = 51/247 (20%)
Query 61 DISWSSAFVGTLFMIICSELGDKTFLITAVLSMRCTKVDSSKKSRPNEKRPGSRWHVFAG 120
DIS+ F+ + +I+ SELGDKT+ I ++SMR SR VF+G
Sbjct 62 DISFYHGFLASFSVIVVSELGDKTWFIAVIMSMRH-----------------SRLTVFSG 104
Query 121 GFLALVVMTLMAAAGG---QVLPLLLKQELRTIFMVCLLVIFGAKMLHEAAVYEENSDEP 177
AL +MT+++A G QV+P + L T L +FG KMLHE N E
Sbjct 105 AMGALALMTVLSACLGWITQVIPRAVTYYLST----ALFALFGLKMLHEGWTMSPN--EG 158
Query 178 EELKDLAQSE----------------------ESMSQSRLFVLARSFFSPVFIQAASLTL 215
+E + AQ+E S S++R L F S +FI+A SLT
Sbjct 159 QEGYEEAQAEVAKREGELDAGKFEMLEGGGGVASQSETRKIFL---FTSRIFIEAFSLTF 215
Query 216 MAEWGDRSQLATFALAADRSASAVCLGAILGHGLCTALAVIGGNVLAKRISERVVLVLGG 275
+AEWGDRSQL T L A + + V G ILGH LCT +AVIGG ++A+RIS R V ++GG
Sbjct 216 VAEWGDRSQLTTIILGARENIAGVIGGGILGHALCTGIAVIGGKIVAQRISVRTVTLIGG 275
Query 276 ICFLSFA 282
+ FL FA
Sbjct 276 VVFLLFA 282
> SPAC17G8.08c
Length=287
Score = 129 bits (324), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 92/256 (35%), Positives = 127/256 (49%), Gaps = 60/256 (23%)
Query 74 MIICSELGDKTFLITAVLSMRCTKVDSSKKSRPNEKRPGSRWHVFAGGFLALVVMTLMAA 133
MI E+GDKTF++ A+L+ SR VFAG + AL +MTL+
Sbjct 56 MIFGCEIGDKTFIVAALLAFE-----------------NSRLTVFAGSYSALFIMTLLGV 98
Query 134 AGGQVLPLLLKQELRTIFMVCLLVIFGAKMLHEAAVYEENSDEPEELKDLAQS------- 186
G PLL ++L I L VIFG KML EA +E D E + D Q+
Sbjct 99 LLGHAAPLLFPRKLTDILGGVLFVIFGIKMLMEA---KEVMDSKESMSDEFQNVRNEIAA 155
Query 187 --------EES-----------------MSQ--SRLFVLARSFFSPVFIQAASLTLMAEW 219
EE MSQ S+ + + FSP+FI+A +LT ++EW
Sbjct 156 NGPIDQLLEEGAAPSHYTGHRSRSGHTLMSQLKSKGRNVMATLFSPLFIKAFALTFVSEW 215
Query 220 GDRSQLATFALAADRSASAVCLGAILGHGLCTALAVIGGNVLAKRISERVVLVLGGICFL 279
GDRSQ+AT A+AA + V +GA +GH CTALAVI G ++ +I V+ +GGI F+
Sbjct 216 GDRSQIATIAMAASDNVYGVFMGANVGHACCTALAVISGKYISTKIKVHKVMFIGGILFI 275
Query 280 SFAAFTAVGELYFHGG 295
+F G +YF+ G
Sbjct 276 AF------GLVYFYQG 285
> 7299965
Length=323
Score = 125 bits (314), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 85/248 (34%), Positives = 129/248 (52%), Gaps = 42/248 (16%)
Query 58 PEGDISWSSAFVGTLFMIICSELGDKTFLITAVLSMRCTKVDSSKKSRPNEKRPGSRWHV 117
P+ ++ AF ++ +I+ +ELGDKTF I A+++MR R V
Sbjct 88 PKTKGTFIDAFTASISVILLTELGDKTFFIAAIMAMR-----------------HPRLIV 130
Query 118 FAGGFLALVVMTLMAAAGGQVLPLLLKQELRTIFM-VCLLVIFGAKMLHEAAVYE-ENSD 175
F G AL +MT+++ A G + K + T ++ L +IFG KML++ Y+ + +D
Sbjct 131 FGGAIAALALMTVLSCAFGMAANFIPK--IYTYYISTALFLIFGLKMLYDG--YKMKPTD 186
Query 176 EPEELK----DLAQSEESMSQSRLFVL---------------ARSFFSPVFIQAASLTLM 216
EEL+ DL + E+ + + L A F + QA ++T +
Sbjct 187 AQEELEEVQTDLRKREDELDRDVNAALVNDAESGRRRPQKRGATYFTMRILAQAFTMTFL 246
Query 217 AEWGDRSQLATFALAADRSASAVCLGAILGHGLCTALAVIGGNVLAKRISERVVLVLGGI 276
AEWGDRSQL T LAA + V G I+GH +CT LAVIGG ++A +IS R V ++GGI
Sbjct 247 AEWGDRSQLTTIILAASKDVYGVIAGGIIGHCICTGLAVIGGRLVASKISVRTVTIVGGI 306
Query 277 CFLSFAAF 284
F+ FA +
Sbjct 307 VFIGFAIY 314
> At1g25520
Length=230
Score = 118 bits (296), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 80/235 (34%), Positives = 113/235 (48%), Gaps = 40/235 (17%)
Query 67 AFVGTLFMIICSELGDKTFLITAVLSMRCTKVDSSKKSRPNEKRPGSRWHVFAGGFLALV 126
F +L M SE+GDKTF A+L+MR R V AG AL+
Sbjct 7 GFTKSLAMTFVSEIGDKTFFAAAILAMRY-----------------PRRLVLAGCLSALI 49
Query 127 VMTLMAAAGGQVLPLLLKQELRTIFMVCLLVIFGAKMLHEA------------------- 167
VMT+++A G P L+ ++ L FG L +
Sbjct 50 VMTILSATLGWAAPNLISRKWTHHITTLLFFGFGLWSLWDGFKEGGGGSEELAEVEAELD 109
Query 168 AVYEENSDEPEELKDLAQSEESMSQSRLFVLARSFFSPVFIQAASLTLMAEWGDRSQLAT 227
A + N P++ + +E+ Q+R F+ FFSP+F++A S+ EWGD+SQLAT
Sbjct 110 ADLKANGKSPKDSSK--REDENKKQNRAFL--TQFFSPIFLKAFSINFFGEWGDKSQLAT 165
Query 228 FALAADRSASAVCLGAILGHGLCTALAVIGGNVLAKRISERVVLVLGGICFLSFA 282
LAAD + V LG ++ LCT AVIGG LA +ISER+V + GG+ F+ F
Sbjct 166 IGLAADENPFGVVLGGVVAQFLCTTAAVIGGKSLASQISERIVALSGGMLFIIFG 220
> At1g68650
Length=228
Score = 117 bits (293), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 82/233 (35%), Positives = 115/233 (49%), Gaps = 38/233 (16%)
Query 67 AFVGTLFMIICSELGDKTFLITAVLSMRCTKVDSSKKSRPNEKRPGSRWHVFAGGFLALV 126
F +L M SE+GDKTF A+L+MR R V AG AL+
Sbjct 7 GFTKSLAMTFLSEIGDKTFFAAAILAMRY-----------------PRRLVLAGCLSALI 49
Query 127 VMTLMAAAGGQVLPLLLKQELRTIFMVCLLVIFGAKMLHEAAVYEE-------------- 172
VMT+++A G P L+ ++ L FG L + ++E
Sbjct 50 VMTILSATLGWAAPNLISRKWTHHITTFLFFGFGLWSLWDG--FKEGGGSEELAEVEAEL 107
Query 173 NSD---EPEELKDLAQSEESMSQSRLFVLARSFFSPVFIQAASLTLMAEWGDRSQLATFA 229
+SD ++ K+ +E Q R F+ A FFSP+F++A S+ EWGD+SQLAT
Sbjct 108 DSDLKKTNDQSKNSKIEDEQKKQKRPFLTA--FFSPIFLKAFSINFFGEWGDKSQLATIG 165
Query 230 LAADRSASAVCLGAILGHGLCTALAVIGGNVLAKRISERVVLVLGGICFLSFA 282
LAAD + V LG I+ LCT AV+GG LA +ISER+V + GG+ F+ F
Sbjct 166 LAADENPLGVVLGGIVAQTLCTTAAVLGGKSLASQISERIVALSGGMLFIIFG 218
> At5g36290
Length=325
Score = 112 bits (280), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 68/202 (33%), Positives = 114/202 (56%), Gaps = 25/202 (12%)
Query 74 MIICSELGDKTFLITAVLSMRCTKVDSSKKSRPNEKRPGSRWHVFAGGFLALVVMTLMAA 133
MI+ +E+GD+TF+I A+++MR K V +G AL VMT+++
Sbjct 91 MILVTEIGDETFIIAALMAMRHPKAT-----------------VLSGALSALFVMTILST 133
Query 134 AGGQVLPLLLKQELRTIFMVCLLVIFGAKMLHEAAVYEENSDEPEELKDLAQSEESMSQS 193
G+++P L+ ++ L FG ++L+ A ++ ++ + + + Q
Sbjct 134 GLGRIVPNLISRKHTNSAATVLYAFFGLRLLYIAWRSTDSKSNQKKEMEEVEEKLESGQG 193
Query 194 -----RLFVLARSFFSPVFIQAASLTLMAEWGDRSQLATFALAADRSASAVCLGAILGHG 248
RLF F +P+F+++ LT +AEWGDRSQ+AT ALA ++A V +GA +GH
Sbjct 194 KTPFRRLF---SRFCTPIFLESFILTFLAEWGDRSQIATIALATHKNAIGVAIGASIGHT 250
Query 249 LCTALAVIGGNVLAKRISERVV 270
+CT+LAV+GG++LA RIS+R +
Sbjct 251 VCTSLAVVGGSMLASRISQRTL 272
> SPAC186.05c
Length=262
Score = 103 bits (258), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 74/239 (30%), Positives = 114/239 (47%), Gaps = 47/239 (19%)
Query 74 MIICSELGDKTFLITAVLSMRCTKVDSSKKSRPNEKRPGSRWHVFAGGFLALVVMTLMAA 133
MII ELGDK+F++TA+L+ + R VF G +LAL MT A
Sbjct 33 MIIGCELGDKSFIVTALLAYQY-----------------GRASVFFGSYLALFFMTSFAV 75
Query 134 AGGQVLPLLLKQELRTIFMVCLLVIFGAKMLHEAAVYEENS----DEPEELKDLAQSEES 189
G+ P L + + I L +IFG KML E+ E+ +E ++++ + +EE
Sbjct 76 LVGRAAPFLFPKSITHILGGTLFLIFGVKMLKESKEVRESQQSLENEFDKVEKIIVNEED 135
Query 190 MSQS--------------------RLFVLA------RSFFSPVFIQAASLTLMAEWGDRS 223
M ++ ++F ++ FS FI+A +L ++E GDRS
Sbjct 136 MKKTLELGLPASNRSSSTLKDKFFKVFSMSCFKNLFSKKFSRAFIKAFALIFVSELGDRS 195
Query 224 QLATFALAADRSASAVCLGAILGHGLCTALAVIGGNVLAKRISERVVLVLGGICFLSFA 282
Q+AT ++A V +G +GH LCT +AVI G ++ +I VL GGI F+ F
Sbjct 196 QIATIVMSAKEKVLDVFIGVNIGHMLCTMVAVIVGRYISNKIEMYKVLFFGGIVFMIFG 254
> YBR187w
Length=280
Score = 101 bits (251), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 75/256 (29%), Positives = 119/256 (46%), Gaps = 46/256 (17%)
Query 56 PVPEGDISWSSAFVGTLFMIICSELGDKTFLITAVLSMRCTKVDSSKKSRPNEKRPGSRW 115
+ G S +F+ ++ MI SE+GDKTFLI A+++MR +V
Sbjct 30 SMESGSSSHLKSFLMSVSMIGLSEIGDKTFLIAALMAMRHKRV----------------- 72
Query 116 HVFAGGFLALVVMTLMAAAGGQVLPLLLKQELRTIFMVCLLVIFGAKMLHEA-------- 167
VF+ +L +MT+++ G L + F L ++FG K+ E
Sbjct 73 LVFSAAATSLAIMTILSGVVGHSAVAFLSERYTAFFAGILFLVFGYKLTMEGLEMSKDAG 132
Query 168 ------------AVYEENSD--EPEELKDLAQSEESMSQS-------RLFVLARSFFSPV 206
A+ + N D + E+ D A ++ + S R+ LA FSPV
Sbjct 133 VEEEMAEVEEEIAIKDMNQDMDDVEKGGDTAYDKQLKNASIGKKIVHRIRELASFMFSPV 192
Query 207 FIQAASLTLMAEWGDRSQLATFALAADRSASAVCLGAILGHGLCTALAVIGGNVLAKRIS 266
++Q + + E GDRSQ++ A+A D V GA++GH +C+ LAV+GG +LA RIS
Sbjct 193 WVQIFLMVFLGELGDRSQISIIAMATDSDYWYVIAGAVIGHAICSGLAVVGGKLLATRIS 252
Query 267 ERVVLVLGGICFLSFA 282
R + + + F FA
Sbjct 253 IRTITLASSLLFFIFA 268
> At4g13590
Length=273
Score = 90.5 bits (223), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 72/217 (33%), Positives = 110/217 (50%), Gaps = 33/217 (15%)
Query 66 SAFVGTLFMIICSELGDKTFLITAVLSMRCTKVDSSKKSRPNEKRPGSRWHVFAGGFLAL 125
S F +I SE+GDKTF I A+L+M+ K V G AL
Sbjct 78 SGFTAAFSLIFVSEIGDKTFFIAALLAMQYEKT-----------------LVLLGSMGAL 120
Query 126 VVMTLMAAAGGQVLPLLLKQELRTIFM-----VCLLVIFGAKMLHEA----AVYEENSDE 176
+MT+++ G++ + Q T+ + + LL+ FG K + +A V +N +E
Sbjct 121 SLMTILSVVIGKIFQSVPAQFQTTLPIGEYAAIALLMFFGLKSIKDAWDLPPVEAKNGEE 180
Query 177 PE-ELKDLAQSEESMSQSRLFVLARSFFSPVFI--QAASLTLMAEWGDRSQLATFALAAD 233
EL + +++EE + + ++ +P+ I ++ SL AEWGDRS LAT AL A
Sbjct 181 TGIELGEYSEAEELVKEKA----SKKLTNPLEILWKSFSLVFFAEWGDRSMLATVALGAA 236
Query 234 RSASAVCLGAILGHGLCTALAVIGGNVLAKRISERVV 270
+S V GAI GH + T LA++GG LA ISE++V
Sbjct 237 QSPLGVASGAIAGHLVATVLAIMGGAFLANYISEKLV 273
> CE27005
Length=255
Score = 85.9 bits (211), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 71/210 (33%), Positives = 98/210 (46%), Gaps = 51/210 (24%)
Query 48 KASAGQVLPVPEGDISWSSAFVGTLFMIICSELGDKTFLITAVLSMRCTKVDSSKKSRPN 107
K + + + DIS+ F+ + +I+ SELGDKT+ I ++SMR
Sbjct 49 KEAQKEHKKISSEDISFYHGFLASFSVIVVSELGDKTWFIAVIMSMRH------------ 96
Query 108 EKRPGSRWHVFAGGFLALVVMTLMAAAGG---QVLPLLLKQELRTIFMVCLLVIFGAKML 164
SR VF+G AL +MT+++A G QV+P + L T L +FG KML
Sbjct 97 -----SRLTVFSGAMGALALMTVLSACLGWITQVIPRAVTYYLST----ALFALFGLKML 147
Query 165 HEAAVYEENSDEPEELKDLAQSE----------------------ESMSQSRLFVLARSF 202
HE N E +E + AQ+E S S++R L F
Sbjct 148 HEGWTMSPN--EGQEGYEEAQAEVAKREGELDAGKFEMLEGGGGVASQSETRKIFL---F 202
Query 203 FSPVFIQAASLTLMAEWGDRSQLATFALAA 232
S +FI+A SLT +AEWGDRSQL T L A
Sbjct 203 TSRIFIEAFSLTFVAEWGDRSQLTTIILGA 232
> At1g64150
Length=398
Score = 85.9 bits (211), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 88/264 (33%), Positives = 119/264 (45%), Gaps = 62/264 (23%)
Query 60 GDISWSSAFVGTLFMIICSELGDKTFLITAVLSMRCTKVDSSKKSRPNEKRPGSRWHVFA 119
GDIS S F +I SELGDKTF I A+L+ R S VF
Sbjct 153 GDIS--SGFASAFLLIFFSELGDKTFFIAALLAAR-----------------NSAATVFV 193
Query 120 GGFLALVVMTLMAAAGG-------QVLPLLL---KQELRTIFMVCLL------------- 156
G F AL +MT+++ G +VLP + I VCLL
Sbjct 194 GTFGALGIMTIISVVLGRTFHYVDEVLPFRFGGTDLPIDDIAAVCLLALLGLDFYVEKVT 253
Query 157 ---------------VIFGAKMLHEAAVYEENSDEPEELKDLAQSEESMSQSRLFVLARS 201
V FG L +A E +DE ++ +LA SE S + + + A +
Sbjct 254 GLTMKMLIGFQAWFMVYFGVSTLLDAVSDEGKADEEQKEAELAVSELSGNGAGIVAAANT 313
Query 202 FFSPVFIQAASLTLMAEWGDRSQLATFALAADRSASAVCLGAILGHGLCTALAVIGGNVL 261
I +L +AEWGD+S +T ALAA S V GA+ GHG T LAV+GG++L
Sbjct 314 -----IISTFALVFVAEWGDKSFFSTIALAAASSPLGVIAGALAGHGAATLLAVLGGSLL 368
Query 262 AKRISERVVLVLGGICFLSFAAFT 285
+SE+ + +GG+ FL FAA T
Sbjct 369 GNFLSEKAIAYVGGVLFLVFAAVT 392
> CE24528
Length=349
Score = 32.0 bits (71), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 21/78 (26%), Positives = 36/78 (46%), Gaps = 15/78 (19%)
Query 184 AQSEESMSQSRLFVLARSF-----FSPVFIQAASLTLMAEWGDRSQLATFALAADRSASA 238
+QS ++ LF +A F F +F+ AA + ++ +Q A+
Sbjct 244 SQSPATLRLQLLFFIAMCFQLSIPFLVIFLPAAYIVFAIQYDYYNQ----------GANN 293
Query 239 VCLGAILGHGLCTALAVI 256
+C+ AI HG+CT L +I
Sbjct 294 LCMAAITFHGVCTTLVMI 311
> At4g35800
Length=1840
Score = 31.6 bits (70), Expect = 1.9, Method: Composition-based stats.
Identities = 16/46 (34%), Positives = 28/46 (60%), Gaps = 1/46 (2%)
Query 156 LVIFGAKMLHEAAVYEENSDEPEELKDLAQSEESMSQSR-LFVLAR 200
L I G KM+ E + + +DEP++L + A+ ++++ R L VL R
Sbjct 180 LTIEGMKMIAEYKIQRKKNDEPDQLPEPAERKQTLGADRVLSVLKR 225
> CE06793
Length=355
Score = 30.0 bits (66), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 26/110 (23%), Positives = 47/110 (42%), Gaps = 23/110 (20%)
Query 59 EGDISWSSAFVGTLFMIICSELGDKTFLITAVLSMRCTKVDSSKKSRPNEKRPGSRWHVF 118
E I+W+ AFVG II + T ++ +L + C ++ + PG R +VF
Sbjct 120 ESYINWTKAFVG----IINDYVSINTMVLLPML-LYCGRIAT---------LPGDRINVF 165
Query 119 AGGFLALV---------VMTLMAAAGGQVLPLLLKQELRTIFMVCLLVIF 159
+ L+ ++ +A G + +L R + ++C L IF
Sbjct 166 PTNMICLILQIVPFFICILNYVATLNGAAVLKVLAATSRVVTLICFLAIF 215
Lambda K H
0.324 0.135 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6743279066
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40