bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2245_orf2
Length=154
Score E
Sequences producing significant alignments: (Bits) Value
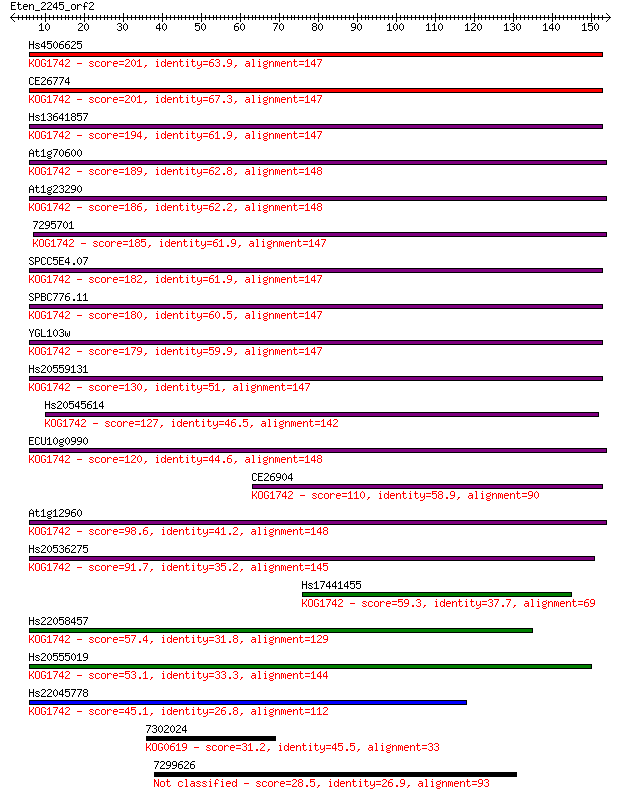
Hs4506625 201 3e-52
CE26774 201 6e-52
Hs13641857 194 7e-50
At1g70600 189 2e-48
At1g23290 186 1e-47
7295701 185 2e-47
SPCC5E4.07 182 2e-46
SPBC776.11 180 8e-46
YGL103w 179 3e-45
Hs20559131 130 1e-30
Hs20545614 127 1e-29
ECU10g0990 120 8e-28
CE26904 110 1e-24
At1g12960 98.6 4e-21
Hs20536275 91.7 5e-19
Hs17441455 59.3 3e-09
Hs22058457 57.4 1e-08
Hs20555019 53.1 2e-07
Hs22045778 45.1 6e-05
7302024 31.2 0.83
7299626 28.5 5.1
> Hs4506625
Length=148
Score = 201 bits (512), Expect = 3e-52, Method: Compositional matrix adjust.
Identities = 94/147 (63%), Positives = 118/147 (80%), Gaps = 1/147 (0%)
Query 6 MTTRLRKTRKLRGHVSAGHGRVGKHRKHPGGRGKAGGMHHKRIHFDKFHPGYFGKVGMRH 65
M +RLRKTRKLRGHVS GHGR+GKHRKHPGGRG AGG+HH RI+FDK+HPGYFGKVGM+H
Sbjct 1 MPSRLRKTRKLRGHVSHGHGRIGKHRKHPGGRGNAGGLHHHRINFDKYHPGYFGKVGMKH 60
Query 66 FHLLRAQQHCPVINVDRLWSLLSPKAFERASSGAAGEKAYVVDCTQAGYFKVLGKGALPK 125
+HL R Q CP +N+D+LW+L+S + A+ G A ++D ++GY+KVLGKG LPK
Sbjct 61 YHLKRNQSFCPTVNLDKLWTLVSEQTRVNAAKNKTGA-APIIDVVRSGYYKVLGKGKLPK 119
Query 126 LPLIVRARFFSKLAEKKIKEAGGVCVL 152
P+IV+A+FFS+ AE+KIK GG CVL
Sbjct 120 QPVIVKAKFFSRRAEEKIKSVGGACVL 146
> CE26774
Length=145
Score = 201 bits (510), Expect = 6e-52, Method: Compositional matrix adjust.
Identities = 99/147 (67%), Positives = 118/147 (80%), Gaps = 4/147 (2%)
Query 6 MTTRLRKTRKLRGHVSAGHGRVGKHRKHPGGRGKAGGMHHKRIHFDKFHPGYFGKVGMRH 65
M LRKTRKLRGHVS GHGR+GKHRKHPGGRG AGG HH RI+ DK+HPGYFGKVGMR
Sbjct 1 MAHALRKTRKLRGHVSHGHGRIGKHRKHPGGRGNAGGQHHHRINRDKYHPGYFGKVGMRV 60
Query 66 FHLLRAQQHCPVINVDRLWSLLSPKAFERASSGAAGEKAYVVDCTQAGYFKVLGKGALPK 125
FHL + Q +CP +NV+RLWSL+ + ++A+ G K+ V+DCT+ GYFKVLGKG LP+
Sbjct 61 FHLNKNQHYCPTVNVERLWSLVPQEVRDKATGG----KSPVIDCTKLGYFKVLGKGLLPE 116
Query 126 LPLIVRARFFSKLAEKKIKEAGGVCVL 152
PLIV+ARFFS AE+KIK+AGG CVL
Sbjct 117 TPLIVKARFFSHEAEQKIKKAGGACVL 143
> Hs13641857
Length=148
Score = 194 bits (492), Expect = 7e-50, Method: Compositional matrix adjust.
Identities = 91/147 (61%), Positives = 115/147 (78%), Gaps = 1/147 (0%)
Query 6 MTTRLRKTRKLRGHVSAGHGRVGKHRKHPGGRGKAGGMHHKRIHFDKFHPGYFGKVGMRH 65
M +RLRKTRKLRGHVS HGR+GKHRKHPGGRG AGG+HH RI+FDK+HPGYFGKVGM+H
Sbjct 1 MPSRLRKTRKLRGHVSHRHGRIGKHRKHPGGRGNAGGLHHHRINFDKYHPGYFGKVGMKH 60
Query 66 FHLLRAQQHCPVINVDRLWSLLSPKAFERASSGAAGEKAYVVDCTQAGYFKVLGKGALPK 125
+HL R Q CP +N+D+LW+L+S + A+ G A ++D ++GY+KVLGKG LPK
Sbjct 61 YHLKRNQSFCPTVNLDKLWTLVSEQTRVNAAKNKTGA-APIIDVVRSGYYKVLGKGKLPK 119
Query 126 LPLIVRARFFSKLAEKKIKEAGGVCVL 152
+ V+A+FFS+ AE+KIK GG CVL
Sbjct 120 QAVTVQAKFFSRRAEEKIKSVGGACVL 146
> At1g70600
Length=146
Score = 189 bits (480), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 93/149 (62%), Positives = 114/149 (76%), Gaps = 5/149 (3%)
Query 6 MTTRLRKTRKLRGHVSAGHGRVGKHRKHPGGRGKAGGMHHKRIHFDKFHPGYFGKVGMRH 65
MTTR +K RK RGHVSAGHGR+GKHRKHPGGRG AGGMHH RI FDK+HPGYFGKVGMR+
Sbjct 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
Query 66 FHLLRAQQHCPVINVDRLWSLLSPKAFERASSGAAGEKAYVVDCTQAGYFKVLGKGALPK 125
FH LR + CP++N+D+LWSL+ E + + + ++D TQ G+FKVLGKG LP+
Sbjct 61 FHKLRNKFFCPIVNLDKLWSLVP----EDVKAKSTKDNVPLIDVTQHGFFKVLGKGHLPE 116
Query 126 L-PLIVRARFFSKLAEKKIKEAGGVCVLS 153
P +V+A+ SK AEKKIKEAGG VL+
Sbjct 117 NKPFVVKAKLISKTAEKKIKEAGGAVVLT 145
> At1g23290
Length=146
Score = 186 bits (473), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 92/149 (61%), Positives = 114/149 (76%), Gaps = 5/149 (3%)
Query 6 MTTRLRKTRKLRGHVSAGHGRVGKHRKHPGGRGKAGGMHHKRIHFDKFHPGYFGKVGMRH 65
M T L+K RK RGHVSAGHGR+GKHRKHPGGRG AGGMHH RI FDK+HPGYFGKVGMR+
Sbjct 1 MATALKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
Query 66 FHLLRAQQHCPVINVDRLWSLLSPKAFERASSGAAGEKAYVVDCTQAGYFKVLGKGALPK 125
FH LR + CP++N+D+LWSL+ E + ++ + ++D TQ G+FKVLGKG LP+
Sbjct 61 FHKLRNKFFCPIVNLDKLWSLVP----EDVKAKSSKDNVPLIDVTQHGFFKVLGKGHLPE 116
Query 126 L-PLIVRARFFSKLAEKKIKEAGGVCVLS 153
P +V+A+ SK AEKKIKEAGG VL+
Sbjct 117 NKPFVVKAKLISKTAEKKIKEAGGAVVLT 145
> 7295701
Length=148
Score = 185 bits (470), Expect = 2e-47, Method: Compositional matrix adjust.
Identities = 91/147 (61%), Positives = 114/147 (77%), Gaps = 1/147 (0%)
Query 7 TTRLRKTRKLRGHVSAGHGRVGKHRKHPGGRGKAGGMHHKRIHFDKFHPGYFGKVGMRHF 66
+ +KTRKLRGHVS GHGR+GKHRKHPGGRG AGGMHH RI+FDK+HPGYFGKVGMR+F
Sbjct 2 NIKRKKTRKLRGHVSHGHGRIGKHRKHPGGRGNAGGMHHHRINFDKYHPGYFGKVGMRNF 61
Query 67 HLLRAQQHCPVINVDRLWSLLSPKAFERASSGAAGEKAYVVDCTQAGYFKVLGKGALPKL 126
HL R + P IN+D+LWSL+ + F + KA V+D + GY+K+LG+G LP
Sbjct 62 HLRRQHKFRPEINLDKLWSLVGAEKFAELEKEKS-TKAPVIDLVKFGYYKLLGRGHLPAR 120
Query 127 PLIVRARFFSKLAEKKIKEAGGVCVLS 153
P+IV+A++FSK AE KIK+AGGVC+LS
Sbjct 121 PVIVKAKYFSKKAEDKIKKAGGVCLLS 147
> SPCC5E4.07
Length=148
Score = 182 bits (463), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 91/147 (61%), Positives = 113/147 (76%), Gaps = 1/147 (0%)
Query 6 MTTRLRKTRKLRGHVSAGHGRVGKHRKHPGGRGKAGGMHHKRIHFDKFHPGYFGKVGMRH 65
M T + KTRKLRGHVSAGHGR+GKHRKHPGGRGKAGG+ H R HFDK+HPGYFGKVGMR
Sbjct 1 MPTHVSKTRKLRGHVSAGHGRIGKHRKHPGGRGKAGGLQHLRSHFDKYHPGYFGKVGMRR 60
Query 66 FHLLRAQQHCPVINVDRLWSLLSPKAFERASSGAAGEKAYVVDCTQAGYFKVLGKGALPK 125
FHL++ P +N+DRLW+LL +A ++ G E A V++ Q+GY KVLGKG LP+
Sbjct 61 FHLMKNPLWRPTVNLDRLWTLLPNEARDK-YLGKNTEVAPVINVLQSGYGKVLGKGRLPE 119
Query 126 LPLIVRARFFSKLAEKKIKEAGGVCVL 152
P+IV+ R+ S+ AE+KIK+AGGV L
Sbjct 120 TPVIVQTRYVSRRAEEKIKQAGGVVEL 146
> SPBC776.11
Length=148
Score = 180 bits (457), Expect = 8e-46, Method: Compositional matrix adjust.
Identities = 89/147 (60%), Positives = 108/147 (73%), Gaps = 1/147 (0%)
Query 6 MTTRLRKTRKLRGHVSAGHGRVGKHRKHPGGRGKAGGMHHKRIHFDKFHPGYFGKVGMRH 65
M T KTRKLRGHVSAGHGR+GKHRKHPGGRGKAGG+ H R HFDK+HPGYFGKVGMR
Sbjct 1 MPTHTSKTRKLRGHVSAGHGRIGKHRKHPGGRGKAGGLQHLRSHFDKYHPGYFGKVGMRR 60
Query 66 FHLLRAQQHCPVINVDRLWSLLSPKAFERASSGAAGEKAYVVDCTQAGYFKVLGKGALPK 125
FHL++ P +N+DRLW+L+ P G E A V++ Q+GY KVLGKG LP
Sbjct 61 FHLMKNPLWRPTVNLDRLWTLV-PNETREKYLGKNTEVAPVINVLQSGYGKVLGKGRLPD 119
Query 126 LPLIVRARFFSKLAEKKIKEAGGVCVL 152
P+I++ R+ S+ AE+KIK+AGGV L
Sbjct 120 TPVIIQTRYVSRRAEEKIKQAGGVVEL 146
> YGL103w
Length=149
Score = 179 bits (453), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 88/147 (59%), Positives = 110/147 (74%), Gaps = 0/147 (0%)
Query 6 MTTRLRKTRKLRGHVSAGHGRVGKHRKHPGGRGKAGGMHHKRIHFDKFHPGYFGKVGMRH 65
M +R KTRK RGHVSAG GR+GKHRKHPGGRG AGG HH RI+ DK+HPGYFGKVGMR+
Sbjct 1 MPSRFTKTRKHRGHVSAGKGRIGKHRKHPGGRGMAGGQHHHRINMDKYHPGYFGKVGMRY 60
Query 66 FHLLRAQQHCPVINVDRLWSLLSPKAFERASSGAAGEKAYVVDCTQAGYFKVLGKGALPK 125
FH +A PV+N+D+LW+L+ ++ A+ E A V+D AGY K+LGKG +P
Sbjct 61 FHKQQAHFWKPVLNLDKLWTLIPEDKRDQYLKSASKETAPVIDTLAAGYGKILGKGRIPN 120
Query 126 LPLIVRARFFSKLAEKKIKEAGGVCVL 152
+P+IV+ARF SKLAE+KI+ AGGV L
Sbjct 121 VPVIVKARFVSKLAEEKIRAAGGVVEL 147
> Hs20559131
Length=123
Score = 130 bits (327), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 75/147 (51%), Positives = 94/147 (63%), Gaps = 26/147 (17%)
Query 6 MTTRLRKTRKLRGHVSAGHGRVGKHRKHPGGRGKAGGMHHKRIHFDKFHPGYFGKVGMRH 65
M +RLRKT+KLRGHVS GHGR+GK +KHP G AGGMHH RI+F+K++PGYFGKVGM
Sbjct 1 MPSRLRKTQKLRGHVSHGHGRIGKLQKHPRGHSNAGGMHHHRINFNKYYPGYFGKVGMSE 60
Query 66 FHLLRAQQHCPVINVDRLWSLLSPKAFERASSGAAGEKAYVVDCTQAGYFKVLGKGALPK 125
+ A ++ P GAA ++D Q+GY+KVLGK LPK
Sbjct 61 QTQVNAAKNKP---------------------GAAP----LIDVVQSGYYKVLGKEKLPK 95
Query 126 LPLIVRARFFSKLAEKKIKEAGGVCVL 152
P+IV+A+FFS+ AE KIK G CVL
Sbjct 96 QPVIVKAKFFSRRAE-KIKGVKGTCVL 121
> Hs20545614
Length=167
Score = 127 bits (318), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 66/148 (44%), Positives = 91/148 (61%), Gaps = 16/148 (10%)
Query 10 LRKTRKLRGHVSAGHGRVGKHRKHPGGRGKAGGMHHKRIHFDKFHPGYFGKVGMRHFHLL 69
LRKTRKL+GHV H + + + P R F+K+HPGYFGKVG H+HL
Sbjct 17 LRKTRKLQGHVIHSHDPIREAPEAPW---------RPRTSFNKYHPGYFGKVGRTHYHLK 67
Query 70 RAQQHCPVINVDRLWSLLSPKAFERASSGAAGEKAYVVDCTQAGYFKVLGKGALPKLPLI 129
R Q CP +N+D+LW+L+S + + A+ G A ++D GY+KVLGKG LPK P+I
Sbjct 68 RNQSFCPTVNLDKLWTLVSEQTWANAAENKTGA-APIIDVALLGYYKVLGKGKLPKQPVI 126
Query 130 VRARFFSKLAEKKIK------EAGGVCV 151
++A+FFS+ AE+K+K GG CV
Sbjct 127 MKAKFFSRRAEEKMKGVAGDLRPGGCCV 154
> ECU10g0990
Length=147
Score = 120 bits (302), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 66/148 (44%), Positives = 93/148 (62%), Gaps = 2/148 (1%)
Query 6 MTTRLRKTRKLRGHVSAGHGRVGKHRKHPGGRGKAGGMHHKRIHFDKFHPGYFGKVGMRH 65
MT R++KTRKLRGHVS G+GRVGKHRKH GGRG AGG H + F +FHP Y GK GMR
Sbjct 1 MTDRVKKTRKLRGHVSHGYGRVGKHRKHSGGRGLAGGFSHMKTFFTRFHPDYHGKRGMRV 60
Query 66 FHLLRAQQHCPVINVDRLWSLLSPKAFERASSGAAGEKAYVVDCTQAGYFKVLGKGALPK 125
+H + I+ RLW ++ PK +R EK V+D + GY V+G +
Sbjct 61 YHRKENSDYARPISSARLWGMI-PKE-QRYDFLDNPEKVPVIDVREFGYHVVVGGKLSLE 118
Query 126 LPLIVRARFFSKLAEKKIKEAGGVCVLS 153
P++V+AR+F+ A+++I + GG +++
Sbjct 119 RPIVVKARYFTPSAKEEITKVGGKWIIT 146
> CE26904
Length=88
Score = 110 bits (274), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 53/90 (58%), Positives = 69/90 (76%), Gaps = 4/90 (4%)
Query 63 MRHFHLLRAQQHCPVINVDRLWSLLSPKAFERASSGAAGEKAYVVDCTQAGYFKVLGKGA 122
MR FHL + Q +CP +NV+RLWSL+ + ++A+ G K+ V+DCT+ GYFKVLGKG
Sbjct 1 MRVFHLNKNQHYCPTVNVERLWSLVPQEVRDKATGG----KSPVIDCTKLGYFKVLGKGL 56
Query 123 LPKLPLIVRARFFSKLAEKKIKEAGGVCVL 152
LP+ PLIV+ARFFS AE+KIK+AGG CVL
Sbjct 57 LPETPLIVKARFFSHEAEQKIKKAGGACVL 86
> At1g12960
Length=104
Score = 98.6 bits (244), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 61/149 (40%), Positives = 76/149 (51%), Gaps = 50/149 (33%)
Query 6 MTTRLRKTRKLRGHVSAGHGRVGKHRKHPGGRGKAGGMHHKRIHFDKFHPGYFGKVGMRH 65
MTT +KTR LR HVS GHGR GKHRK PG RG AG VGMR+
Sbjct 1 MTTSRKKTRNLREHVSVGHGRFGKHRKLPGSRGNAG-------------------VGMRY 41
Query 66 FHLLRAQQHCPVINVDRLWSLLSPKAFERASSGAAGEKAYVVDCTQAGYFKVLGKGALPK 125
FH LR + +C ++N+D+LWS+ VLGKG LP+
Sbjct 42 FHKLRNKFYCQIVNLDKLWSM------------------------------VLGKGFLPE 71
Query 126 L-PLIVRARFFSKLAEKKIKEAGGVCVLS 153
P++V+A+ S EKKIKEAG VL+
Sbjct 72 NKPVVVKAKLVSNTDEKKIKEAGSAVVLT 100
> Hs20536275
Length=166
Score = 91.7 bits (226), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 51/145 (35%), Positives = 74/145 (51%), Gaps = 33/145 (22%)
Query 6 MTTRLRKTRKLRGHVSAGHGRVGKHRKHPGGRGKAGGMHHKRIHFDKFHPGYFGKVGMRH 65
M +RLRKT+KL G+VS GH K +I+FDK+HPGYFGKVGMRH
Sbjct 1 MPSRLRKTQKLLGYVSHGHSCTVK-----------------KINFDKYHPGYFGKVGMRH 43
Query 66 FHLLRAQQHCPVINVDRLWSLLSPKAFERASSGAAGEKAYVVDCTQAGYFKVLGKGALPK 125
+HL R Q CP +N+D+LW+L+ + + + + G A +VD +
Sbjct 44 YHLKRNQSFCPAVNLDKLWTLVREQTWVKVAKNKTGA-APIVDVS--------------- 87
Query 126 LPLIVRARFFSKLAEKKIKEAGGVC 150
L L++ F+ K E+ +C
Sbjct 88 LFLVIVILFYVTWVRSKYPESTEIC 112
> Hs17441455
Length=127
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 45/69 (65%), Gaps = 1/69 (1%)
Query 76 PVINVDRLWSLLSPKAFERASSGAAGEKAYVVDCTQAGYFKVLGKGALPKLPLIVRARFF 135
P +N+D+LW+L+ + A+ G A + D +GY++VLG+ P P+IV+A+FF
Sbjct 41 PAVNLDKLWALVRKQTQVNAAKNKPGT-APITDVVPSGYYRVLGREKFPNQPVIVKAKFF 99
Query 136 SKLAEKKIK 144
S+ A++KI+
Sbjct 100 SRRAQEKIQ 108
> Hs22058457
Length=101
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 41/130 (31%), Positives = 59/130 (45%), Gaps = 30/130 (23%)
Query 6 MTTRLRKTRKLRGHVSAGHGRVGKHRKHPGGRGKAGGMHHKRIHFDKFHPGYFGKVGMRH 65
M +RLRK+RKL+GH S GHG H G +A M G
Sbjct 1 MPSRLRKSRKLQGHTSHGHG----HITECGSTVEAVVMQ-----------VVCTTTGSTL 45
Query 66 FHLL-RAQQHCPVINVDRLWSLLSPKAFERASSGAAGEKAYVVDCTQAGYFKVLGKGALP 124
++ R Q P +N D+LW+++ + + Q GY+KVLGKG LP
Sbjct 46 TNVTQRKQSFSPTVNFDKLWTMI--------------QAPPITAVVQLGYYKVLGKGKLP 91
Query 125 KLPLIVRARF 134
K +I++A+F
Sbjct 92 KQSVIMKAQF 101
> Hs20555019
Length=137
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 48/148 (32%), Positives = 68/148 (45%), Gaps = 59/148 (39%)
Query 6 MTTRLRKTRKLRGHVSAGHGRVGKHRKHPGGRGKAGGMHHKRIHFDKFHPGYFGKVGMRH 65
M + +RKT+KL GH+S GH + KH+KH GG + H+R+
Sbjct 1 MPSGMRKTQKLWGHMSHGHCLISKHQKHSGGMSR-----HRRM----------------- 38
Query 66 FHLLRAQQHCPVINVDRLWSLLSPKAFERASSGAAGEKAYVVDCTQAGYFKVLGKGALPK 125
LL+ + I+V VD Y+KVLGKG LPK
Sbjct 39 --LLKTRLGLSTIDV--------------------------VD-----YYKVLGKGKLPK 65
Query 126 LPLIVRARFFSKLAEKKIK----EAGGV 149
P+IV+A+FF + A+++IK AGGV
Sbjct 66 HPIIVKAKFFRRRAQERIKGCWSGAGGV 93
> Hs22045778
Length=236
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 30/116 (25%), Positives = 49/116 (42%), Gaps = 7/116 (6%)
Query 6 MTTRLRKTRKLRGHVSAGHGRVGKHRKHPGGRGKAGGMHHKRIHFDKFHPGYFGKVGMRH 65
M +R RK L GH+S R + K PGG G + + P + +
Sbjct 1 MPSRRRKIWNLWGHMSLLLHR--QTLKLPGGPGSCWDHASPQDQLPQITPRFLWESWYAA 58
Query 66 FHLLRAQQ----HCPVINVDRLWSLLSPKAFERASSGAAGEKAYVVDCTQAGYFKV 117
L + CP +N D+LW+L+S + + A+ G ++D G++KV
Sbjct 59 LPLEVEPELCPNDCPTVNFDKLWTLVSEQTWVNAAKTKTG-IGPIIDVVWPGFYKV 113
> 7302024
Length=1315
Score = 31.2 bits (69), Expect = 0.83, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 21/43 (48%), Gaps = 10/43 (23%)
Query 36 GRGKAGGMHHKRI--------HFDKFHPGYF--GKVGMRHFHL 68
GR + GGM+H I + HPGYF ++ + H HL
Sbjct 697 GRNEHGGMYHSNIKILDLSHNNISIIHPGYFRPAEISLTHLHL 739
> 7299626
Length=338
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 25/93 (26%), Positives = 36/93 (38%), Gaps = 15/93 (16%)
Query 38 GKAGGMHHKRIHFDKFHPGYFGKVGMRHFHLLRAQQHCPVINVDRLWSLLSPKAFERASS 97
GK G H + + F K L+ Q HC +NVD S + ++ E S
Sbjct 40 GKTYGEHKEYLEFSKDQ-------------YLQLQAHCKELNVDFTASAMDERSLEFLS- 85
Query 98 GAAGEKAYVVDCTQAGYFKVLGKGALPKLPLIV 130
A + A F +L K A LPL++
Sbjct 86 -ALNVPFIKIGSGDANNFPLLKKAANLNLPLVI 117
Lambda K H
0.325 0.141 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1997677330
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40