bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2249_orf1
Length=223
Score E
Sequences producing significant alignments: (Bits) Value
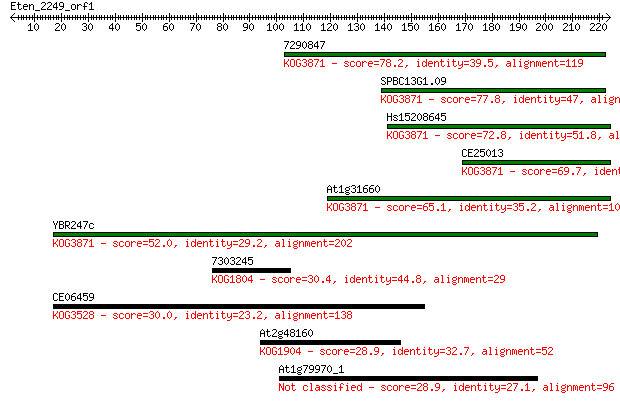
7290847 78.2 1e-14
SPBC13G1.09 77.8 2e-14
Hs15208645 72.8 6e-13
CE25013 69.7 5e-12
At1g31660 65.1 1e-10
YBR247c 52.0 9e-07
7303245 30.4 2.9
CE06459 30.0 3.5
At2g48160 28.9 7.7
At1g79970_1 28.9 8.6
> 7290847
Length=436
Score = 78.2 bits (191), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 47/124 (37%), Positives = 73/124 (58%), Gaps = 8/124 (6%)
Query 103 DEETADKDGFVVLDCEEDEEDLVFEKQRTGSSATRATVNLADLVMEQLKK-----QTENK 157
DEE + D LD +ED+ FE+ + + + T++L+ ++M+++++ T+
Sbjct 90 DEEVNETDLMADLDMDEDDVA-AFERFQQPAQEGKRTLHLSKMIMQKIQEKEADIHTKIS 148
Query 158 DKSQEKRVESSLSPKVVEVYTAMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTMW 217
D+ K E + PKV E+Y + L RYRSGK+PKAFKIIP+L WE+IL +T P W
Sbjct 149 DEGSLKIEE--IDPKVKEMYEGVRDVLKRYRSGKIPKAFKIIPKLRNWEQILFITEPHNW 206
Query 218 SKQA 221
S A
Sbjct 207 SAAA 210
> SPBC13G1.09
Length=449
Score = 77.8 bits (190), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 39/92 (42%), Positives = 60/92 (65%), Gaps = 12/92 (13%)
Query 139 TVNLADLVMEQLKKQTENKDKSQEKRVESS---------LSPKVVEVYTAMAPFLARYRS 189
T +L+DL+M+++ E + +++ + + S+ L PKV+EVY+ + L++YRS
Sbjct 154 TTSLSDLIMQKI---NEAEARARGEYIPSAEEEENALPPLPPKVIEVYSKVGVLLSKYRS 210
Query 190 GKMPKAFKIIPRLHAWEEILVLTNPTMWSKQA 221
GK+PKAFKIIP L WE+IL LT P MW+ A
Sbjct 211 GKIPKAFKIIPTLSNWEDILYLTRPDMWTPHA 242
> Hs15208645
Length=301
Score = 72.8 bits (177), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 43/90 (47%), Positives = 57/90 (63%), Gaps = 7/90 (7%)
Query 141 NLADLVMEQL-KKQTENKDKSQEKR--VESSLSPKVVEVYTAMAPFLARYRSGKMPKAFK 197
LAD++ME+L +KQTE + E L P+V+EVY + L++YRSGK+PKAFK
Sbjct 10 TLADIIMEKLTEKQTEVETVMSEVSGFPMPQLDPRVLEVYRGVREVLSKYRSGKLPKAFK 69
Query 198 IIPRLHAWEEILVLTNPTMWS----KQATR 223
IIP L WE+IL +T P W+ QATR
Sbjct 70 IIPALSNWEQILYVTEPEAWTAAAMYQATR 99
> CE25013
Length=449
Score = 69.7 bits (169), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 30/59 (50%), Positives = 43/59 (72%), Gaps = 4/59 (6%)
Query 169 LSPKVVEVYTAMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTMWS----KQATR 223
+ P+VVE+Y + ++++YRSGK+PKAFKIIP++ WE+IL LT P W+ QATR
Sbjct 179 MDPEVVEMYEQIGQYMSKYRSGKVPKAFKIIPKMINWEQILFLTKPETWTAAAMYQATR 237
> At1g31660
Length=442
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 65/109 (59%), Gaps = 9/109 (8%)
Query 119 EDEEDLVFEKQRTGSSATRATVNLADLVMEQLKKQTENKDKSQEKRVESSLSPKVVEVYT 178
ED+E L FE ++ + T L D+++++LK + + D ++E+R + + P + ++Y
Sbjct 118 EDDEKL-FESFLNKNAPPQRT--LTDIIIKKLKDK--DADLAEEERPDPKMDPAITKLYK 172
Query 179 AMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTMWS----KQATR 223
+ F++ Y GK+PKAFK++ + WE++L LT P WS QATR
Sbjct 173 GVGKFMSEYTVGKLPKAFKLVTSMEHWEDVLYLTEPEKWSPNALYQATR 221
> YBR247c
Length=483
Score = 52.0 bits (123), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 59/258 (22%), Positives = 103/258 (39%), Gaps = 71/258 (27%)
Query 17 RHNPLYEDVLDARGIPLPGGAIDAPRNVKIKQAVEDAEAVENCTRDDQEQEGLLPPKLGK 76
RH+PL +D+ A+G + + K+ ++ A + E++G + K +
Sbjct 13 RHDPLLKDLDAAQG---------TLKKINKKKLAQNDAANHDAA---NEEDGYIDSKASR 60
Query 77 KLLKLVEEQQ---RAEGIA---------------------------------DASDAEGG 100
K+L+L +EQQ E +A D SD E
Sbjct 61 KILQLAKEQQDEIEGEELAESERNKQFEARFTTMSYDDEDEDEDEDEEAFGEDISDFEPE 120
Query 101 GPDEETADKDGFVVLDCEEDEEDLVFEKQRTGSSATRATVNLADLVMEQLKK---QTE-- 155
G +E + D E+ + K+ ++ + NLAD +M +++ Q E
Sbjct 121 GDYKEEEEIVEIDEEDAAMFEQ---YFKKSDDFNSLSGSYNLADKIMASIREKESQVEDM 177
Query 156 -------NKDKSQEKRVESSLSP--------KVVEVYTAMAPFLARYRSGKMPKAFKIIP 200
N+ + + S L KV++ YT + L + GK+PK FK+IP
Sbjct 178 QDDEPLANEQNTSRGNISSGLKSGEGVALPEKVIKAYTTVGSILKTWTHGKLPKLFKVIP 237
Query 201 RLHAWEEILVLTNPTMWS 218
L W++++ +TNP WS
Sbjct 238 SLRNWQDVIYVTNPEEWS 255
> 7303245
Length=1194
Score = 30.4 bits (67), Expect = 2.9, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 76 KKLLKLVEEQQRAEGIADASDAEGGGPDE 104
K+ L++EQ R GIA +D GG P E
Sbjct 5 KEKFNLLDEQHRLRGIASTTDVPGGAPAE 33
> CE06459
Length=703
Score = 30.0 bits (66), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 32/142 (22%), Positives = 59/142 (41%), Gaps = 27/142 (19%)
Query 17 RHNPLYEDVLDARGIPLPGGAIDAPRNVKIKQAVEDAEAVENCTRDDQEQEGLLPPKLGK 76
R++PL+ P G I P ++ A + CT D +Q+ LPP+ +
Sbjct 332 RYHPLFRSNTLTHVHFNPLGTISTPTSI--------ATTPQRCTSTDSKQKHALPPQ--R 381
Query 77 KLLKLVEEQQRAEGIADASDAEGGGPDEETADKDGFVVLDCEEDEEDLVFEKQRTGSSAT 136
++++ + A+ + A DA+ D DG + DC E T S T
Sbjct 382 EIIE-PDSTTCADDNSQARDADNSRMDSSDYGSDG--ISDCRES----------TASDGT 428
Query 137 RAT----VNLADLVMEQLKKQT 154
+ T + + D++ +++K T
Sbjct 429 QKTLSDRIEVTDIIFDKVKSPT 450
> At2g48160
Length=1191
Score = 28.9 bits (63), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 94 ASDAEGGGPDEETADKDGFVVLDCEEDEEDLVFEKQRTGSSATRATVNLADL 145
SD+EGG E +D DG + E + E+ + S+A R T+ L D+
Sbjct 858 GSDSEGGCDSEGGSDSDGGDFESVTPEHESRILEENVSSSTAERHTLILEDV 909
> At1g79970_1
Length=263
Score = 28.9 bits (63), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 26/99 (26%), Positives = 44/99 (44%), Gaps = 6/99 (6%)
Query 101 GPDEETADKDGFVVLDCEEDE---EDLVFEKQRTGSSATRATVNLADLVMEQLKKQTENK 157
GP+ +T G +LDC+ + ED+ ++ S +T N ++ E K++EN
Sbjct 163 GPNPKTTYIFGDCILDCDPKDLGKEDIEIHEEGDDSFSTEKPQNHREVSAE---KESENA 219
Query 158 DKSQEKRVESSLSPKVVEVYTAMAPFLARYRSGKMPKAF 196
+ E L P + + +A L YRSG + F
Sbjct 220 GAEESCYEEEDLFPMAMPLNPNLAESLENYRSGVVAAMF 258
Lambda K H
0.311 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4255059914
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40