bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2255_orf1
Length=110
Score E
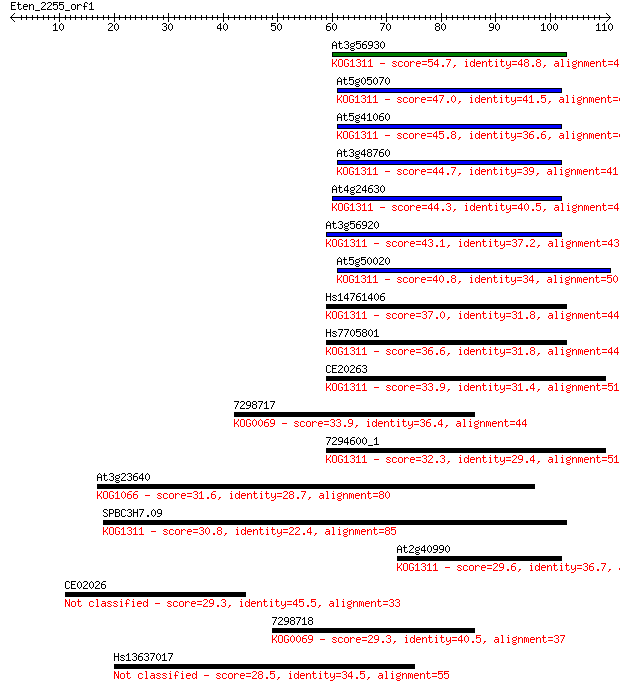
Sequences producing significant alignments: (Bits) Value
At3g56930 54.7 4e-08
At5g05070 47.0 9e-06
At5g41060 45.8 2e-05
At3g48760 44.7 4e-05
At4g24630 44.3 5e-05
At3g56920 43.1 1e-04
At5g50020 40.8 6e-04
Hs14761406 37.0 0.010
Hs7705801 36.6 0.010
CE20263 33.9 0.071
7298717 33.9 0.076
7294600_1 32.3 0.23
At3g23640 31.6 0.39
SPBC3H7.09 30.8 0.67
At2g40990 29.6 1.4
CE02026 29.3 1.7
7298718 29.3 1.8
Hs13637017 28.5 2.8
> At3g56930
Length=477
Score = 54.7 bits (130), Expect = 4e-08, Method: Composition-based stats.
Identities = 21/43 (48%), Positives = 31/43 (72%), Gaps = 0/43 (0%)
Query 60 QRLYESWEGENIFCCGGRLMTGPEPLHLAVSVLLLLGPCVVYY 102
+RLY+ W G N F CGGRL+ GP+ L +S +L+LGP V+++
Sbjct 9 KRLYQVWRGSNKFLCGGRLIFGPDASSLYLSTILILGPAVMFF 51
> At5g05070
Length=339
Score = 47.0 bits (110), Expect = 9e-06, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 27/41 (65%), Gaps = 0/41 (0%)
Query 61 RLYESWEGENIFCCGGRLMTGPEPLHLAVSVLLLLGPCVVY 101
R Y++W+G N FCCGGRL+ GP+ L ++ L+ P + +
Sbjct 37 RFYKAWKGNNRFCCGGRLIFGPDVSSLYLTSFLIGAPALTF 77
> At5g41060
Length=410
Score = 45.8 bits (107), Expect = 2e-05, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 28/41 (68%), Gaps = 0/41 (0%)
Query 61 RLYESWEGENIFCCGGRLMTGPEPLHLAVSVLLLLGPCVVY 101
R+Y++W+G NIFC GR + GP+ L +++ L++ P ++
Sbjct 19 RVYQTWKGSNIFCLQGRFIFGPDVRSLGLTISLIVAPVTIF 59
> At3g48760
Length=470
Score = 44.7 bits (104), Expect = 4e-05, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 27/41 (65%), Gaps = 0/41 (0%)
Query 61 RLYESWEGENIFCCGGRLMTGPEPLHLAVSVLLLLGPCVVY 101
R Y+ W+G N+F GGRL+ GP+ + ++V L+ P +V+
Sbjct 28 RTYKGWKGNNVFFLGGRLVFGPDARSILITVFLITAPVIVF 68
> At4g24630
Length=374
Score = 44.3 bits (103), Expect = 5e-05, Method: Composition-based stats.
Identities = 17/42 (40%), Positives = 30/42 (71%), Gaps = 0/42 (0%)
Query 60 QRLYESWEGENIFCCGGRLMTGPEPLHLAVSVLLLLGPCVVY 101
QR+++ W+G N F GGRL+ GP+ L +++LL++ P V++
Sbjct 3 QRVFQVWKGSNKFILGGRLIFGPDARSLPLTLLLIIVPVVLF 44
> At3g56920
Length=319
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 28/43 (65%), Gaps = 0/43 (0%)
Query 59 QQRLYESWEGENIFCCGGRLMTGPEPLHLAVSVLLLLGPCVVY 101
+QR+Y+ W +N F CGGRL+ GP+ L ++ ++ GP + +
Sbjct 8 RQRIYQVWPAKNKFYCGGRLVFGPDASSLLLTTCMIGGPAIAF 50
> At5g50020
Length=414
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 29/50 (58%), Gaps = 0/50 (0%)
Query 61 RLYESWEGENIFCCGGRLMTGPEPLHLAVSVLLLLGPCVVYYQVVMPLIR 110
R++E+W+G N F GGRL+ GP+ + + LL++ P + V +R
Sbjct 4 RVFEAWKGSNKFLFGGRLIFGPDAWSIPFTFLLIITPVCFFSVFVATHLR 53
> Hs14761406
Length=382
Score = 37.0 bits (84), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 27/47 (57%), Gaps = 3/47 (6%)
Query 59 QQRLYESWE---GENIFCCGGRLMTGPEPLHLAVSVLLLLGPCVVYY 102
++++ WE G N FCC GR+M + +++ L+LG C +++
Sbjct 25 RKKVTRKWEKLPGRNTFCCDGRVMMARQKGIFYLTLFLILGTCTLFF 71
> Hs7705801
Length=382
Score = 36.6 bits (83), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 27/47 (57%), Gaps = 3/47 (6%)
Query 59 QQRLYESWE---GENIFCCGGRLMTGPEPLHLAVSVLLLLGPCVVYY 102
++++ WE G N FCC GR+M + +++ L+LG C +++
Sbjct 25 RKKVTRKWEKLPGRNTFCCDGRVMMARQKGIFYLTLFLILGTCTLFF 71
> CE20263
Length=368
Score = 33.9 bits (76), Expect = 0.071, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 59 QQRLYESWEGENIFCCGGRLMTGPEPLHLAVSVLLLLGPCVVYYQVVMPLI 109
++R ++S G N F CGGRL+ V+V+L++ VY+ P +
Sbjct 37 KRRKWQSHPGRNRFGCGGRLVCSRSHGAFVVTVILMIATLTVYFVFDAPFL 87
> 7298717
Length=248
Score = 33.9 bits (76), Expect = 0.076, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 24/44 (54%), Gaps = 1/44 (2%)
Query 42 LPQLLLEVPLQRGRLFGQQRLYESWEGENIFCCGGRLMTGPEPL 85
+ Q + V + RG++ Q LYE+ + IF G + T PEPL
Sbjct 155 MKQTAVLVNIARGKIVNQDDLYEALKANRIFSAGLDV-TDPEPL 197
> 7294600_1
Length=260
Score = 32.3 bits (72), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 15/54 (27%), Positives = 25/54 (46%), Gaps = 3/54 (5%)
Query 59 QQRLYESWE---GENIFCCGGRLMTGPEPLHLAVSVLLLLGPCVVYYQVVMPLI 109
QR+ WE G N F C G LM+ P ++ +L+ G +++ P +
Sbjct 15 NQRVTRKWELFAGRNKFYCDGLLMSAPHTGVFYLTCILITGTSALFFAFDCPFL 68
> At3g23640
Length=365
Score = 31.6 bits (70), Expect = 0.39, Method: Composition-based stats.
Identities = 23/82 (28%), Positives = 36/82 (43%), Gaps = 23/82 (28%)
Query 17 GILMAQPEVEST--GDEAHRPQLQQRTLPQLLLEVPLQRGRLFGQQRLYESWEGENIFCC 74
G+LMA+ E D+ RP + L R G QR +W G+N
Sbjct 192 GMLMARSTYEGMELADKNKRPFV-------------LTRAGFIGSQRYAATWTGDN---- 234
Query 75 GGRLMTGPEPLHLAVSVLLLLG 96
++ E LH+++S++L LG
Sbjct 235 ----LSNWEHLHMSISMVLQLG 252
> SPBC3H7.09
Length=350
Score = 30.8 bits (68), Expect = 0.67, Method: Composition-based stats.
Identities = 19/85 (22%), Positives = 40/85 (47%), Gaps = 7/85 (8%)
Query 18 ILMAQPEVESTGDEAHRPQLQQRTLPQLLLEVPLQRGRLFGQQRLYESWEGENIFCCGGR 77
+ ++ P +E ++ + +++P + EVP R Y++ G NI+ C GR
Sbjct 25 VTLSDPTYPMNLEEKNQIPYRFQSVPDDVPEVPHIESR-------YKNLPGNNIYLCCGR 77
Query 78 LMTGPEPLHLAVSVLLLLGPCVVYY 102
L + +S+ L+ P V+++
Sbjct 78 LQMSSQYKAFLISLFALILPGVLFF 102
> At2g40990
Length=340
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 11/30 (36%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 72 FCCGGRLMTGPEPLHLAVSVLLLLGPCVVY 101
F CGGRL+ GP+ L ++ ++ GP + +
Sbjct 43 FYCGGRLVFGPDASSLLLTTAMIGGPALTF 72
> CE02026
Length=401
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 24/43 (55%), Gaps = 10/43 (23%)
Query 11 LHCPLKGILMAQPEVESTGDEAHR----------PQLQQRTLP 43
LH L+GIL+++P+ ++G +HR QL+ R LP
Sbjct 32 LHPSLRGILLSRPKRWNSGSPSHRIAVNLVRKYKKQLKPRVLP 74
> 7298718
Length=598
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 21/37 (56%), Gaps = 1/37 (2%)
Query 49 VPLQRGRLFGQQRLYESWEGENIFCCGGRLMTGPEPL 85
V + RG++ Q LYE+ + IF G +M PEPL
Sbjct 512 VNVGRGKIVNQDDLYEALKSNRIFAAGLDVMD-PEPL 547
> Hs13637017
Length=333
Score = 28.5 bits (62), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 26/55 (47%), Gaps = 1/55 (1%)
Query 20 MAQPEVESTGDEAHRPQLQQRTLPQLLLEVPLQRGRLFGQQRLYESWEGENIFCC 74
MA+ V G R L + +L + L+ G L+GQ RLY W E +F C
Sbjct 134 MAKISVAEDGRLRIRGALMGTYVASVLCKSVLEAGFLYGQWRLY-GWTMEPVFVC 187
Lambda K H
0.323 0.143 0.448
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1195973986
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40