bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2259_orf2
Length=369
Score E
Sequences producing significant alignments: (Bits) Value
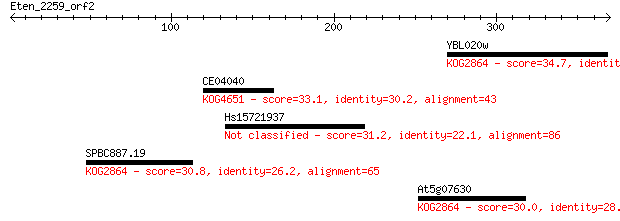
YBL020w 34.7 0.37
CE04040 33.1 0.95
Hs15721937 31.2 3.6
SPBC887.19 30.8 4.3
At5g07630 30.0 8.4
> YBL020w
Length=574
Score = 34.7 bits (78), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 23/112 (20%), Positives = 50/112 (44%), Gaps = 21/112 (18%)
Query 270 CVFGPSTVA-----------SSSGCVLTLQVYCCYVGCLSLFGLLDAYAAATSSGTSL-- 316
VFGP+ + S++ + T++VYC Y+ LSL G+ +A+ + ++G +
Sbjct 382 IVFGPANSSFLLQFLIGSKWSTTSVLDTIRVYCFYIPFLSLNGIFEAFFQSVATGDQILK 441
Query 317 -ELLQRCYSGATAAHLILMPLSVRFAVVYGVPSGAGAAAAQVVGTIIRISCC 367
+SG + L+ ++ ++ G + ++ ++RI C
Sbjct 442 HSYFMMAFSGIFLLNSWLLIEKLKLSI-------EGLILSNIINMVLRILYC 486
> CE04040
Length=321
Score = 33.1 bits (74), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 13/43 (30%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 120 RLFPSSSMAECKKQHVPAAGKQVEKEPFCTRRKTCSPKSQKGE 162
RLF ++M + +P K + + C+RR TC+P +++ E
Sbjct 55 RLFALNAMVKANSHTLPPRSKNISAKNLCSRRMTCAPHNRRYE 97
> Hs15721937
Length=167
Score = 31.2 bits (69), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 19/86 (22%), Positives = 33/86 (38%), Gaps = 4/86 (4%)
Query 133 QHVPAAGKQVEKEPFCTRRKTCSPKSQKGEREQHIDYRHQNELAQFYCVERKELLESDSN 192
+ +P A + + P C + + + KG++ + H A Y RK L +
Sbjct 19 RELPCAWRALHTSPVCAKNRAARVRVSKGDKPVTYEEAH----APHYIAHRKGWLSLHTG 74
Query 193 RRTGTDRGGAGRLESLFLGRLMTLLF 218
G D +E +FL + M F
Sbjct 75 NLDGEDHAAERTVEDVFLRKFMWGTF 100
> SPBC887.19
Length=527
Score = 30.8 bits (68), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 17/65 (26%), Positives = 29/65 (44%), Gaps = 0/65 (0%)
Query 48 KLVLQNGEQLLLLLLLDSKAAAEYALVSGTASVICRVVFAPTESAAFDSFRVLQAQHSAA 107
K ++ G+++++ A YAL S S++ R+VF P E + F L +
Sbjct 255 KHLITKGDKIMVAWYASPSAQGPYALASNYGSLLARIVFRPVEDHSHIVFAQLTHYKNKK 314
Query 108 DRAAA 112
D A
Sbjct 315 DEKKA 319
> At5g07630
Length=401
Score = 30.0 bits (66), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 31/66 (46%), Gaps = 2/66 (3%)
Query 252 LGLASAAAGVLFAAPALRCVFGPSTVASSSGCVLTLQVYCCYVGCLSLFGLLDAYAAATS 311
+GL A G ++ +R ++G S L LQ YC Y+ L++ G +A+ A
Sbjct 235 IGLIFMAFGPSYSYSLIRLLYGEK--WSDGEASLALQFYCLYIIVLAMNGTSEAFLHAVG 292
Query 312 SGTSLE 317
+ LE
Sbjct 293 TKNELE 298
Lambda K H
0.323 0.133 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9085535074
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40