bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2273_orf1
Length=130
Score E
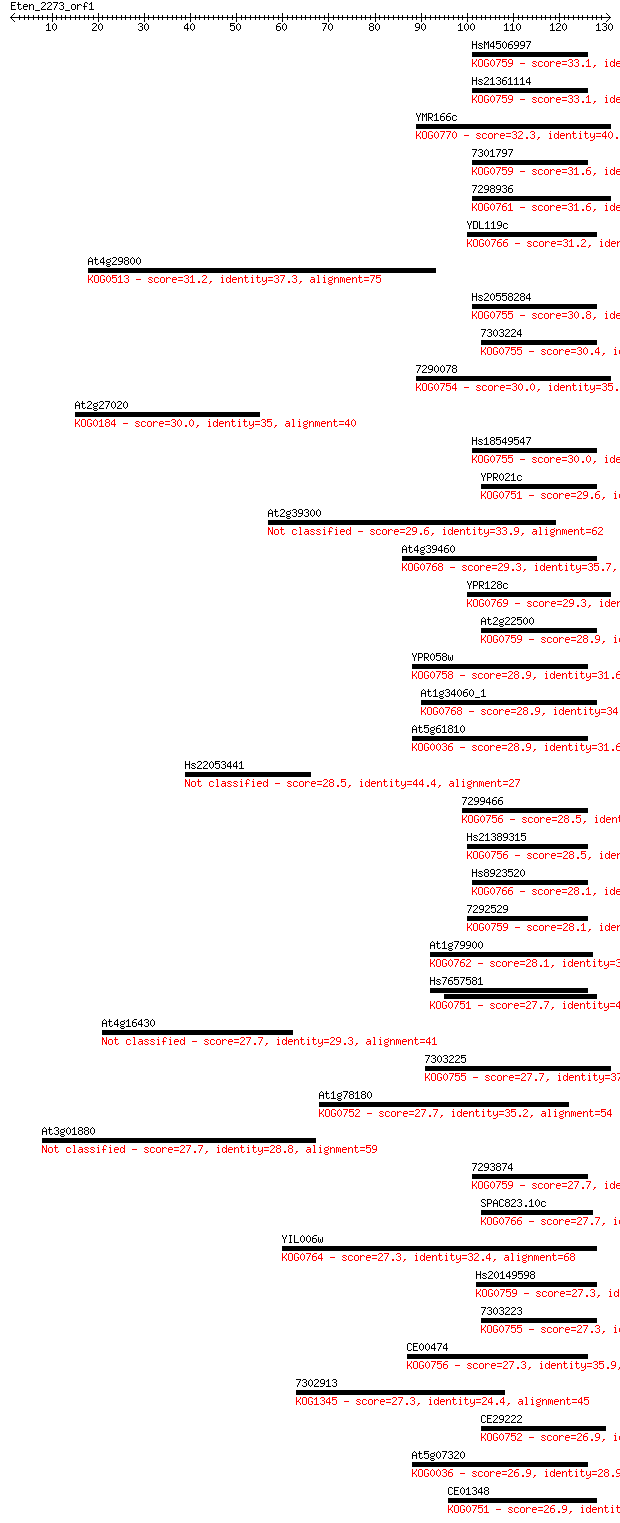
Sequences producing significant alignments: (Bits) Value
HsM4506997 33.1 0.15
Hs21361114 33.1 0.15
YMR166c 32.3 0.21
7301797 31.6 0.35
7298936 31.6 0.45
YDL119c 31.2 0.55
At4g29800 31.2 0.58
Hs20558284 30.8 0.59
7303224 30.4 0.81
7290078 30.0 1.0
At2g27020 30.0 1.1
Hs18549547 30.0 1.1
YPR021c 29.6 1.3
At2g39300 29.6 1.6
At4g39460 29.3 2.1
YPR128c 29.3 2.2
At2g22500 28.9 2.3
YPR058w 28.9 2.3
At1g34060_1 28.9 2.5
At5g61810 28.9 2.5
Hs22053441 28.5 3.3
7299466 28.5 3.7
Hs21389315 28.5 3.8
Hs8923520 28.1 4.0
7292529 28.1 4.2
At1g79900 28.1 4.7
Hs7657581 27.7 5.0
At4g16430 27.7 5.3
7303225 27.7 5.4
At1g78180 27.7 5.5
At3g01880 27.7 5.9
7293874 27.7 6.0
SPAC823.10c 27.7 6.2
YIL006w 27.3 6.7
Hs20149598 27.3 6.7
7303223 27.3 6.7
CE00474 27.3 7.3
7302913 27.3 8.2
CE29222 26.9 8.6
At5g07320 26.9 8.8
CE01348 26.9 9.5
> HsM4506997
Length=314
Score = 33.1 bits (74), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 101 LYGAISSCAAGVFLQPLDIIKTRQQ 125
L+G ++ A VF+QPLD++K R Q
Sbjct 26 LFGGLAGMGATVFVQPLDLVKNRMQ 50
> Hs21361114
Length=314
Score = 33.1 bits (74), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 101 LYGAISSCAAGVFLQPLDIIKTRQQ 125
L+G ++ A VF+QPLD++K R Q
Sbjct 26 LFGGLAGMGATVFVQPLDLVKNRMQ 50
> YMR166c
Length=368
Score = 32.3 bits (72), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 89 GSRGETSSARMGLYGAISSCAAGVFLQPLDIIKTRQQQQCAP 130
G GE S L GA + AG+ P+D++KTR Q Q P
Sbjct 249 GRDGELSIPNEILTGACAGGLAGIITTPMDVVKTRVQTQQPP 290
> 7301797
Length=317
Score = 31.6 bits (70), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 101 LYGAISSCAAGVFLQPLDIIKTRQQ 125
L+G +S A + +QPLD++KTR Q
Sbjct 22 LFGGLSGMGATMVVQPLDLVKTRMQ 46
> 7298936
Length=449
Score = 31.6 bits (70), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 20/34 (58%), Gaps = 4/34 (11%)
Query 101 LYGAISSCAAGV----FLQPLDIIKTRQQQQCAP 130
L IS+C + F+ PLD+IKTR Q Q +P
Sbjct 91 LQQVISACTGAMITACFMTPLDVIKTRMQSQQSP 124
> YDL119c
Length=307
Score = 31.2 bits (69), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 14/28 (50%), Positives = 19/28 (67%), Gaps = 3/28 (10%)
Query 100 GLYGAISSCAAGVFLQPLDIIKTRQQQQ 127
G +G ++S A LQPLD++KTR QQ
Sbjct 17 GFFGGLTSAVA---LQPLDLLKTRIQQD 41
> At4g29800
Length=526
Score = 31.2 bits (69), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 39/79 (49%), Gaps = 5/79 (6%)
Query 18 SYKIFSIQQR--VFSSVHFVRLRISPRPFRPSLAMGGPDPRPTGITEAEAAAPGSDNSTT 75
+Y+IFSI + +F L I P RP + GP PR + +T PG+ +S+
Sbjct 56 NYEIFSILESKFLFGYEDPRLLWIPQSPLRPGDSEAGPSPR-SPLTPNGVVLPGTPSSSF 114
Query 76 RRSRGN--SAAIFGGGSRG 92
R RG +I GGG RG
Sbjct 115 RSPRGRICVLSIDGGGMRG 133
> Hs20558284
Length=300
Score = 30.8 bits (68), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 101 LYGAISSCAAGVFLQPLDIIKTRQQQQ 127
L +++C A VF PL+++KTR Q Q
Sbjct 4 LMSGLAACGACVFTNPLEVVKTRMQLQ 30
> 7303224
Length=304
Score = 30.4 bits (67), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 103 GAISSCAAGVFLQPLDIIKTRQQQQ 127
G +++ AGVF P+++IKTR Q Q
Sbjct 9 GGVAAMGAGVFTNPVEVIKTRIQLQ 33
> 7290078
Length=306
Score = 30.0 bits (66), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 25/46 (54%), Gaps = 4/46 (8%)
Query 89 GSRGETSSARMGLYGAISSCAAGVF----LQPLDIIKTRQQQQCAP 130
G + + S A+ + ++ +AG +QPLD++KTR Q Q P
Sbjct 3 GQQHDISHAKRAAFQVLAGGSAGFLEVCIMQPLDVVKTRIQIQATP 48
> At2g27020
Length=249
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 15 YGNSYKIFSIQQRVFSSVHFVRLRISPRPFRPSLAMGGPD 54
YG++ + + +RV S VH L RPF + +GG D
Sbjct 103 YGDAVPVKELSERVASYVHLCTLYWWLRPFGCGVILGGYD 142
> Hs18549547
Length=304
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 101 LYGAISSCAAGVFLQPLDIIKTRQQQQ 127
+ GA + C A VF PL+++KTR Q Q
Sbjct 11 VLGASACCLACVFTNPLEVVKTRLQLQ 37
> YPR021c
Length=902
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 11/25 (44%), Positives = 15/25 (60%), Gaps = 0/25 (0%)
Query 103 GAISSCAAGVFLQPLDIIKTRQQQQ 127
G+I+ C + P+D IKTR Q Q
Sbjct 537 GSIAGCIGATVVYPIDFIKTRMQAQ 561
> At2g39300
Length=768
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 21/75 (28%), Positives = 34/75 (45%), Gaps = 16/75 (21%)
Query 57 PTGITEAEAAAPGSDNSTTRRSRGNSAAIF-------------GGGSRGETSSARMGLYG 103
P G+ ++++ G+ RRSR S+A F SR T + YG
Sbjct 36 PKGLNKSQSEVSGA---ALRRSRSLSSAAFVIDGTSSNQHRLRNHSSRCLTPERQFKEYG 92
Query 104 AISSCAAGVFLQPLD 118
++S+C++ V Q LD
Sbjct 93 SMSTCSSNVSSQVLD 107
> At4g39460
Length=330
Score = 29.3 bits (64), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 19/42 (45%), Gaps = 0/42 (0%)
Query 86 FGGGSRGETSSARMGLYGAISSCAAGVFLQPLDIIKTRQQQQ 127
+ +R E S L GA + G PLD+IKTR Q
Sbjct 216 YKKAARRELSDPENALIGAFAGALTGAVTTPLDVIKTRLMVQ 257
> YPR128c
Length=328
Score = 29.3 bits (64), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 100 GLYGAISSCAAGVFLQPLDIIKTRQQQQCAP 130
L GA++S A + + PLD+ KT Q Q +P
Sbjct 7 ALTGAVASAMANIAVYPLDLSKTIIQSQVSP 37
> At2g22500
Length=313
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 13/25 (52%), Positives = 15/25 (60%), Gaps = 0/25 (0%)
Query 103 GAISSCAAGVFLQPLDIIKTRQQQQ 127
G I+S AG PLD+IK R Q Q
Sbjct 9 GGIASIVAGCSTHPLDLIKVRMQLQ 33
> YPR058w
Length=307
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 12/38 (31%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 88 GGSRGETSSARMGLYGAISSCAAGVFLQPLDIIKTRQQ 125
G R + + ++ ++GA+S A + + PLD+IK+ Q
Sbjct 211 GLERKDIPAWKLCIFGALSGTALWLMVYPLDVIKSVMQ 248
> At1g34060_1
Length=302
Score = 28.9 bits (63), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 19/38 (50%), Gaps = 0/38 (0%)
Query 90 SRGETSSARMGLYGAISSCAAGVFLQPLDIIKTRQQQQ 127
+R + + + GA + GV PLD+IKTR Q
Sbjct 226 ARRDLNDPENAMIGAFAGAVTGVLTTPLDVIKTRLMVQ 263
> At5g61810
Length=476
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 12/38 (31%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 88 GGSRGETSSARMGLYGAISSCAAGVFLQPLDIIKTRQQ 125
GG+ G+ ++ L G ++ A + P+D++KTR Q
Sbjct 287 GGADGDIGTSGRLLAGGLAGAVAQTAIYPMDLVKTRLQ 324
> Hs22053441
Length=988
Score = 28.5 bits (62), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 39 ISPRPFRPSLAMGGPDPRPTGITEAEA 65
+ P P PSL GP P+P G+ +EA
Sbjct 690 VGPSPQDPSLEASGPSPKPAGVDISEA 716
> 7299466
Length=317
Score = 28.5 bits (62), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 19/27 (70%), Gaps = 0/27 (0%)
Query 99 MGLYGAISSCAAGVFLQPLDIIKTRQQ 125
+G++GAI+ A+ PLD++KTR Q
Sbjct 231 VGVFGAIAGAASVFGNTPLDVVKTRMQ 257
> Hs21389315
Length=311
Score = 28.5 bits (62), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 100 GLYGAISSCAAGVFLQPLDIIKTRQQ 125
G++GAI+ A+ PLD+IKTR Q
Sbjct 224 GVFGAIAGAASVFGNTPLDVIKTRMQ 249
> Hs8923520
Length=304
Score = 28.1 bits (61), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 101 LYGAISSCAAGVFLQPLDIIKTRQQ 125
L G+IS + + QPLD++KTR Q
Sbjct 32 LCGSISGTCSTLLFQPLDLLKTRLQ 56
> 7292529
Length=301
Score = 28.1 bits (61), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 17/26 (65%), Gaps = 4/26 (15%)
Query 100 GLYGAISSCAAGVFLQPLDIIKTRQQ 125
GL G + +C +QPLD++KTR Q
Sbjct 20 GLAGMLGTC----IVQPLDLVKTRMQ 41
> At1g79900
Length=296
Score = 28.1 bits (61), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 92 GETSSARMGLYGAISSCAAGVFLQPLDIIKTRQQQ 126
G+ + M + G ++ A+ V PLD++KTR QQ
Sbjct 196 GQENLRTMLVAGGLAGVASWVACYPLDVVKTRLQQ 230
> Hs7657581
Length=675
Score = 27.7 bits (60), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 92 GETSSARMGLYGAISSCAAGVFLQPLDIIKTRQQ 125
G+ S + L GAI+ A + P D+IKTR Q
Sbjct 516 GQVSPGSLLLAGAIAGMPAASLVTPADVIKTRLQ 549
Score = 27.3 bits (59), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 12/33 (36%), Positives = 20/33 (60%), Gaps = 1/33 (3%)
Query 95 SSARMGLYGAISSCAAGVFLQPLDIIKTRQQQQ 127
S+ R GL G+++ + P+D++KTR Q Q
Sbjct 328 SAYRFGL-GSVAGAVGATAVYPIDLVKTRMQNQ 359
> At4g16430
Length=467
Score = 27.7 bits (60), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 12/41 (29%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 21 IFSIQQRVFSSVHFVRLRISPRPFRPSLAMGGPDPRPTGIT 61
+ + + VF FV+ + +P+ F L++GG PR I
Sbjct 218 VIEMVKSVFGGSDFVQAKEAPKIFGRQLSLGGAKPRSMSIN 258
> 7303225
Length=303
Score = 27.7 bits (60), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 19/40 (47%), Gaps = 0/40 (0%)
Query 91 RGETSSARMGLYGAISSCAAGVFLQPLDIIKTRQQQQCAP 130
+GE S L+GAI F P +IKT+ Q Q A
Sbjct 100 KGEVSYGMGLLWGAIGGVVGCYFSSPFFLIKTQLQSQAAK 139
> At1g78180
Length=262
Score = 27.7 bits (60), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 29/55 (52%), Gaps = 9/55 (16%)
Query 68 PGSDNSTTRRSRGNSAAIFGGGSRGETSSARMGLY-GAISSCAAGVFLQPLDIIK 121
PG DN R R ++ GG R T + R L+ GA+++ + FL PL+ +K
Sbjct 103 PGKDN----RKR----SVIGGVRRRGTMNTRKHLWAGAVAAMVSKTFLAPLERLK 149
> At3g01880
Length=592
Score = 27.7 bits (60), Expect = 5.9, Method: Composition-based stats.
Identities = 17/59 (28%), Positives = 22/59 (37%), Gaps = 15/59 (25%)
Query 8 FWSPLCPYGNSYKIFSIQQRVFSSVHFVRLRISPRPFRPSLAMGGPDPRPTGITEAEAA 66
FW PLCP G H V L ++ P +PSL + +TE A
Sbjct 182 FWQPLCPNG---------------YHAVGLYVTTSPMKPSLGQNSISCVRSDLTEQSEA 225
> 7293874
Length=190
Score = 27.7 bits (60), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 11/25 (44%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 101 LYGAISSCAAGVFLQPLDIIKTRQQ 125
L A++ C AGV P+++I TR Q
Sbjct 24 LVAALAGCVAGVVGTPMELINTRMQ 48
> SPAC823.10c
Length=296
Score = 27.7 bits (60), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 12/24 (50%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 103 GAISSCAAGVFLQPLDIIKTRQQQ 126
GA+ + LQPLD++KTR QQ
Sbjct 21 GALGGFISSTTLQPLDLLKTRCQQ 44
> YIL006w
Length=373
Score = 27.3 bits (59), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 34/72 (47%), Gaps = 11/72 (15%)
Query 60 ITEAEAAAPGSDNSTTRRSRGNSAAIFGGGSRGE----TSSARMGLYGAISSCAAGVFLQ 115
++ A+ P NS+T +I G R + +S+ L GA + +GV +
Sbjct 44 LSSADIIEPIKMNSSTE-------SIIGTTLRKKWVPLSSTQITALSGAFAGFLSGVAVC 96
Query 116 PLDIIKTRQQQQ 127
PLD+ KTR Q Q
Sbjct 97 PLDVAKTRLQAQ 108
> Hs20149598
Length=287
Score = 27.3 bits (59), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 10/26 (38%), Positives = 15/26 (57%), Gaps = 0/26 (0%)
Query 102 YGAISSCAAGVFLQPLDIIKTRQQQQ 127
+G ++SC A PLD++K Q Q
Sbjct 12 FGGLASCGAACCTHPLDLLKVHLQTQ 37
> 7303223
Length=124
Score = 27.3 bits (59), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 11/25 (44%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 103 GAISSCAAGVFLQPLDIIKTRQQQQ 127
G ++ A VF P+D++KTR Q Q
Sbjct 9 GGTAAMGAVVFTNPIDVVKTRMQLQ 33
> CE00474
Length=312
Score = 27.3 bits (59), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 87 GGGSRGETSSARMGLYGAISSCAAGVFLQPLDIIKTRQQ 125
GG + S +GL GA++ A+ P+D++KTR Q
Sbjct 211 GGDNTQPISKPIVGLMGAVAGAASVYGNTPIDVVKTRMQ 249
> 7302913
Length=577
Score = 27.3 bits (59), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 11/45 (24%), Positives = 28/45 (62%), Gaps = 0/45 (0%)
Query 63 AEAAAPGSDNSTTRRSRGNSAAIFGGGSRGETSSARMGLYGAISS 107
+++A+P S+ STT + N+ ++ GSR + ++ + +Y ++ +
Sbjct 437 SKSASPASNYSTTASTLNNADSLMTLGSRQDLLASNLTMYTSMET 481
> CE29222
Length=313
Score = 26.9 bits (58), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 103 GAISSCAAGVFLQPLDIIKTRQQQQCA 129
GA+S C A PLD+I+TR Q A
Sbjct 126 GALSGCLAMTAAMPLDVIRTRLVAQKA 152
> At5g07320
Length=479
Score = 26.9 bits (58), Expect = 8.8, Method: Composition-based stats.
Identities = 11/38 (28%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 88 GGSRGETSSARMGLYGAISSCAAGVFLQPLDIIKTRQQ 125
GG G+ ++ + G ++ A + P+D++KTR Q
Sbjct 288 GGEDGDIGTSGRLMAGGMAGALAQTAIYPMDLVKTRLQ 325
> CE01348
Length=702
Score = 26.9 bits (58), Expect = 9.5, Method: Composition-based stats.
Identities = 11/32 (34%), Positives = 18/32 (56%), Gaps = 0/32 (0%)
Query 96 SARMGLYGAISSCAAGVFLQPLDIIKTRQQQQ 127
SA L G+++ + P+D++KTR Q Q
Sbjct 364 SAYRFLLGSVAGACGATAVYPIDLVKTRMQNQ 395
Lambda K H
0.318 0.133 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1246445644
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40