bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2276_orf1
Length=192
Score E
Sequences producing significant alignments: (Bits) Value
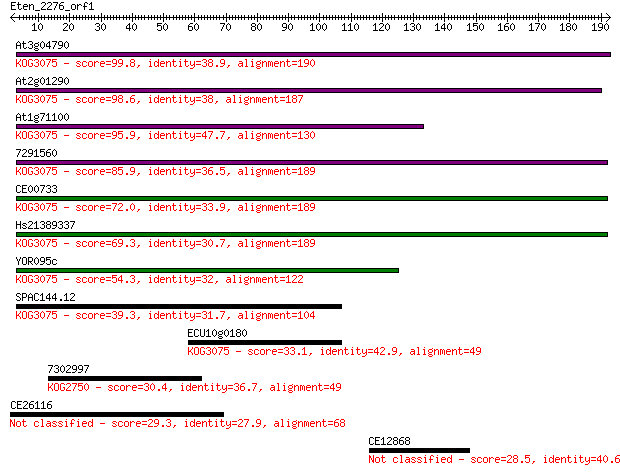
At3g04790 99.8 3e-21
At2g01290 98.6 7e-21
At1g71100 95.9 5e-20
7291560 85.9 5e-17
CE00733 72.0 7e-13
Hs21389337 69.3 4e-12
YOR095c 54.3 1e-07
SPAC144.12 39.3 0.004
ECU10g0180 33.1 0.37
7302997 30.4 2.3
CE26116 29.3 5.2
CE12868 28.5 9.6
> At3g04790
Length=276
Score = 99.8 bits (247), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 74/192 (38%), Positives = 106/192 (55%), Gaps = 30/192 (15%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLP-LDVT 61
LGTGSTA+FAV+++ + + GEL DI TS+ T + A SLGI ++ LD + P +D+
Sbjct 68 LGTGSTAAFAVDQIGKLLSSGELYDIVGIPTSKRTEEQARSLGIPLVGLD--THPRIDLA 125
Query 62 IDFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFGTTG-AVA 120
ID ADEV N + L+KGRGGALLREK+VE + FI VAD+ K+V G +G A+
Sbjct 126 IDGADEVDPN---LDLVKGRGGALLREKMVEAVADKFIVVADDTKLVT--GLGGSGLAMP 180
Query 121 LEVVQFGAQATRQAVLSAVVSTLGGEVLEAEEETPEEAAERLGVSAVFRHHENRKELFVS 180
+EVVQF + ++ + G + R + K +V+
Sbjct 181 VEVVQFCWNFNLIRL--------------------QDLFKEFGCESKLRVDGDGKP-YVT 219
Query 181 DNGNLCLDLFFK 192
DN N +DL+FK
Sbjct 220 DNSNYIIDLYFK 231
> At2g01290
Length=265
Score = 98.6 bits (244), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 71/187 (37%), Positives = 105/187 (56%), Gaps = 26/187 (13%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLPLDVTI 62
LGTGSTA AV+R+ E +RQG+L +I TS+ T++ A SLGI + LD + +D++I
Sbjct 56 LGTGSTAKHAVDRIGELLRQGKLENIVGIPTSKKTQEQALSLGIPLSDLDAHPV-IDLSI 114
Query 63 DFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFGTTGAVALE 122
D ADEV + L+KGRGG+LLREK++E SK F+ + D+ K+V+ G+ A+ +E
Sbjct 115 DGADEV---DPFLNLVKGRGGSLLREKMIEGASKKFVVIVDDSKMVKHIG-GSKLALPVE 170
Query 123 VVQFGAQATRQAVLSAVVSTLGGEVLEAEEETPEEAAERLGVSAVFRHHENRKELFVSDN 182
+V F + T + + S + E G A R E K FV+DN
Sbjct 171 IVPFCWKFTAEKLRSLL--------------------EGYGCEANLRLGEKGKA-FVTDN 209
Query 183 GNLCLDL 189
GN +D+
Sbjct 210 GNYIVDM 216
> At1g71100
Length=267
Score = 95.9 bits (237), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 62/131 (47%), Positives = 84/131 (64%), Gaps = 7/131 (5%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLPLDVTI 62
LGTGSTA AV R++E +R+G+L DI TS T + A SLGI + LD + +D++I
Sbjct 52 LGTGSTAKHAVARISELLREGKLKDIIGIPTSTTTHEQAVSLGIPLSDLDSHPV-VDLSI 110
Query 63 DFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFGTTG-AVAL 121
D ADEV + L+KGRGG+LLREK++E SK F+ + DE K+V+ G +G AV +
Sbjct 111 DGADEV---DPALNLVKGRGGSLLREKMIEGASKKFVVIVDESKLVK--YIGGSGLAVPV 165
Query 122 EVVQFGAQATR 132
EVV F TR
Sbjct 166 EVVPFCCDFTR 176
> 7291560
Length=325
Score = 85.9 bits (211), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 69/195 (35%), Positives = 94/195 (48%), Gaps = 38/195 (19%)
Query 3 LGTGSTASFAVERLAERI-RQGELTDISCASTSEATRKLAESLGINVLSLDEISLPLDVT 61
+G+GST +AV+R+AER+ ++GELTD+ C +S R L +N+ LD + +DV
Sbjct 115 IGSGSTVVYAVQRIAERVWKEGELTDLICVPSSYQARHLILDYNLNLGDLDR-NPNIDVA 173
Query 62 IDFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFGTTGAVAL 121
ID ADEV + MVLIKG GG LL+EK+V +K FI VAD K
Sbjct 174 IDGADEVDR---HMVLIKGGGGCLLQEKVVASCAKHFIVVADYTK--------------- 215
Query 122 EVVQFGAQATRQ-----AVLSAVVSTLGGEVLEAEEETPEEAAERLGVSAVFRHHENRKE 176
++ G Q R A ++ V L E L G A R + +
Sbjct 216 NSIRLGEQWCRGVPIEVAPMAYVPIKLHIEAL-------------FGGEASLRMAKVKAG 262
Query 177 LFVSDNGNLCLDLFF 191
V+DNGN LD F
Sbjct 263 PIVTDNGNFLLDWKF 277
> CE00733
Length=251
Score = 72.0 bits (175), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 64/192 (33%), Positives = 86/192 (44%), Gaps = 34/192 (17%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLP-LDVT 61
+G+GST + VE L + + G L DI C TS T++ G+ V LD S P LDV
Sbjct 38 VGSGSTVKYLVEYLKQGFQNGSLKDIICVPTSFLTKQWLIESGLPVSDLD--SHPELDVC 95
Query 62 IDFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQP--DSFGTTGAV 119
ID ADEV Q IKG GG L +EKIV+ +K F +AD K + D + V
Sbjct 96 IDGADEV---DGQFTCIKGGGGCLAQEKIVQTAAKNFYVIADYLKDSKHLGDRYPN---V 149
Query 120 ALEVVQFGAQATRQAVLSAVVSTLGGEVLEAEEETPEEAAERLGVSAVFRHHENRKELFV 179
+EV+ A Q +L ++ GG S R + V
Sbjct 150 PIEVLPLAA----QPLLRSIPRAEGG-------------------SCQLRQAVKKCGPIV 186
Query 180 SDNGNLCLDLFF 191
+DNGN +D F
Sbjct 187 TDNGNFIIDWQF 198
> Hs21389337
Length=237
Score = 69.3 bits (168), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 58/189 (30%), Positives = 86/189 (45%), Gaps = 28/189 (14%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLPLDVTI 62
+G+GST AV+R+AER++Q L ++ C TS R+L G+ + LD +D+ I
Sbjct 28 IGSGSTIVHAVQRIAERVKQENL-NLVCIPTSFQARQLILQYGLTLSDLDR-HPEIDLAI 85
Query 63 DFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFGTTGAVALE 122
D ADEV + + LIKG GG L +EKIV + FI +AD K + + +E
Sbjct 86 DGADEV---DADLNLIKGGGGCLTQEKIVAGYASRFIVIADFRKDSKNLGDQWHKGIPIE 142
Query 123 VVQFGAQATRQAVLSAVVSTLGGEVLEAEEETPEEAAERLGVSAVFRHHENRKELFVSDN 182
V+ +AV +++ G R N+ V+DN
Sbjct 143 VIPMAYVPVSRAV-----------------------SQKFGGVVELRMAVNKAGPVVTDN 179
Query 183 GNLCLDLFF 191
GN LD F
Sbjct 180 GNFILDWKF 188
> YOR095c
Length=258
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 39/128 (30%), Positives = 65/128 (50%), Gaps = 11/128 (8%)
Query 3 LGTGSTASFAVERLAERIRQGELTDIS----CASTSEATRKLAESLGINVLSLDEISLPL 58
+G+GST + ER+ + + + +++ C T +R L + + S+++ +
Sbjct 44 IGSGSTVVYVAERIGQYLHDPKFYEVASKFICIPTGFQSRNLILDNKLQLGSIEQYPR-I 102
Query 59 DVTIDFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFGTTG- 117
D+ D ADEV +N + LIKG G L +EK+V ++K FI VAD K P G
Sbjct 103 DIAFDGADEVDEN---LQLIKGGGACLFQEKLVSTSAKTFIVVADSRK-KSPKHLGKNWR 158
Query 118 -AVALEVV 124
V +E+V
Sbjct 159 QGVPIEIV 166
> SPAC144.12
Length=274
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 33/105 (31%), Positives = 54/105 (51%), Gaps = 9/105 (8%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLP-LDVT 61
+G+GST + VERL + + + + T +++L + G+ + D P +DV+
Sbjct 38 IGSGSTVVYVVERL---LTKPGVDSVVFIPTGFQSKQLIVNNGLRLGDPD--CYPNVDVS 92
Query 62 IDFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGK 106
D ADEV N + IKG G L +EK++ +K + VAD K
Sbjct 93 FDGADEVDDN---LQCIKGGGACLFQEKLIAFLAKRLVIVADSRK 134
> ECU10g0180
Length=201
Score = 33.1 bits (74), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 21/49 (42%), Positives = 28/49 (57%), Gaps = 3/49 (6%)
Query 58 LDVTIDFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGK 106
LD+ ID AD KNG+ +IKG GGAL EK++ +K I + K
Sbjct 63 LDLYIDGADYFDKNGN---MIKGGGGALTTEKLLYSMAKETIIIVQSHK 108
> 7302997
Length=987
Score = 30.4 bits (67), Expect = 2.3, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 23/49 (46%), Gaps = 0/49 (0%)
Query 13 VERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLPLDVT 61
VE+ ERI Q + DI + S K + I DE+SLP VT
Sbjct 388 VEKKQERIEQSSVMDIVVKNLSSVPPKKESEVAIETEENDEVSLPEVVT 436
> CE26116
Length=618
Score = 29.3 bits (64), Expect = 5.2, Method: Composition-based stats.
Identities = 19/68 (27%), Positives = 34/68 (50%), Gaps = 6/68 (8%)
Query 1 STLGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLPLDV 60
S + T S++ A + ++ R DI+ +S++ R N ++DE L DV
Sbjct 209 SKVETCSSSPQANQEISSEDRNSHHPDIASSSSATTCRS------SNCFNMDEGILTRDV 262
Query 61 TIDFADEV 68
+DF+DE+
Sbjct 263 EMDFSDEI 270
> CE12868
Length=913
Score = 28.5 bits (62), Expect = 9.6, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 116 TGAVALEVVQFGAQATRQAVLSAVVSTLGGEV 147
T A E+++ G+ AT+ + LSA++S L G+V
Sbjct 370 TDQAAKEIMRIGSLATKLSQLSAIISPLAGKV 401
Lambda K H
0.315 0.131 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3238438140
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40