bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2283_orf1
Length=172
Score E
Sequences producing significant alignments: (Bits) Value
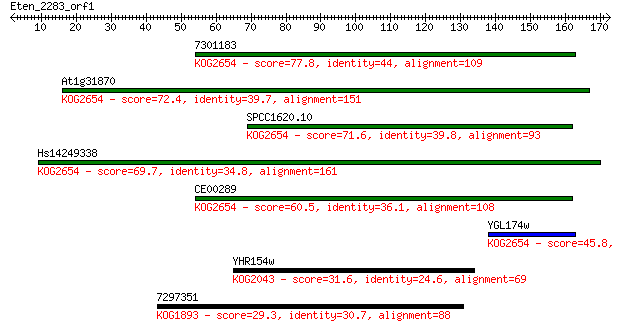
7301183 77.8 1e-14
At1g31870 72.4 4e-13
SPCC1620.10 71.6 8e-13
Hs14249338 69.7 3e-12
CE00289 60.5 2e-09
YGL174w 45.8 4e-05
YHR154w 31.6 0.84
7297351 29.3 4.0
> 7301183
Length=647
Score = 77.8 bits (190), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 48/109 (44%), Positives = 63/109 (57%), Gaps = 9/109 (8%)
Query 54 WGAGLKQKEDLEARKKEEEKIAKQPFARYDIDEEHDAELRARDRWDDPLAKAPSNTAQGL 113
WG GLKQ ED ++R +E A +P ARY DE+ D LR ++ DDP+ +
Sbjct 524 WGKGLKQIEDHKSRLEEMVHEASKPVARYANDEDLDRHLREQEHADDPMLEY-------- 575
Query 114 GRKQKGKEDVDREKKHQRPKCPHDAPPNRFGIKPGYRWDGVVRGNGYEE 162
+QK K+ ++ K PK P NRFGI+PGYRWDGV R NGYE+
Sbjct 576 -MRQKRKKRDQQDNKPVMPKYEGSFPENRFGIRPGYRWDGVDRSNGYEQ 623
> At1g31870
Length=561
Score = 72.4 bits (176), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 60/169 (35%), Positives = 84/169 (49%), Gaps = 30/169 (17%)
Query 16 VIFRDR-EGRIISEEEWLTLEGAKGRKRQRERGPAPVLEWGAGLKQKEDLEARKKEEEKI 74
+FRD+ G+ IS+EE+L + K ++ +E LEWG GL QK + EAR +E E
Sbjct 385 AVFRDKITGKRISKEEYLKSKQKKVIEKPKEIK----LEWGKGLAQKREAEARLQELELE 440
Query 75 AKQPFARYDIDEEHDAELRARDRWDDPLAKAPSNTAQGLGRKQKGKEDV-----DREKKH 129
+PFAR D E D ++ R R+ DP+A L +K+K + + D E K
Sbjct 441 KDKPFARTRDDPELDQMMKERVRFGDPMA--------HLVKKRKYETTLVDLGDDEEMKK 492
Query 130 QRPKCPHDAP------------PNRFGIKPGYRWDGVVRGNGYEEERLK 166
P P NR+GIKPG WDGV R NG E++ +K
Sbjct 493 SGFIIPQSVPKHSWLTRRLEAASNRYGIKPGRHWDGVDRSNGTEKDLIK 541
> SPCC1620.10
Length=305
Score = 71.6 bits (174), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 37/93 (39%), Positives = 50/93 (53%), Gaps = 17/93 (18%)
Query 69 KEEEKIAKQPFARYDIDEEHDAELRARDRWDDPLAKAPSNTAQGLGRKQKGKEDVDREKK 128
KE E+ P ARY+ D E++ EL+ R RW+DP A +N +G
Sbjct 206 KELERQKTVPLARYEDDPEYNKELKERSRWNDPAASFLTNKPVSSKATYQGY-------- 257
Query 129 HQRPKCPHDAPPNRFGIKPGYRWDGVVRGNGYE 161
APPNRF I+PG+RWDG++RGNG+E
Sbjct 258 ---------APPNRFNIRPGHRWDGIIRGNGFE 281
> Hs14249338
Length=619
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 56/164 (34%), Positives = 81/164 (49%), Gaps = 18/164 (10%)
Query 9 GEIPASRVIFRDREGRIISEEEWLTLEGAKGRKRQRERGPAPVL--EWGAGLKQKEDLEA 66
E + +FRD+ GR + L LE + R++ + L +WG GL Q +
Sbjct 454 AEFQYAETVFRDKSGR----KRNLKLERLEQRRKAEKDSERDELYAQWGKGLAQSRQQQQ 509
Query 67 RKKEEEKIAKQPFARYDIDEEHDAELRARDRWDDPLAKAPSNTAQGLGRKQKGKEDVDRE 126
++ K ++P ARY DE+ D LR ++R DP+A N + K+ + V
Sbjct 510 NVEDAMKEMQKPLARYIDDEDLDRMLREQEREGDPMA----NFIKKNKAKENKNKKV--- 562
Query 127 KKHQRPKCPHDAPP-NRFGIKPGYRWDGVVRGNGYEEERLKALC 169
RP+ APP NRF I PGYRWDGV R NG+E++R L
Sbjct 563 ----RPRYSGPAPPPNRFNIWPGYRWDGVDRSNGFEQKRFARLA 602
> CE00289
Length=458
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 39/108 (36%), Positives = 56/108 (51%), Gaps = 9/108 (8%)
Query 54 WGAGLKQKEDLEARKKEEEKIAKQPFARYDIDEEHDAELRARDRWDDPLAKAPSNTAQGL 113
W G+ Q ED A+ +E ++A +P AR D+ +A L+ DP+A +
Sbjct 335 WNKGVAQIEDRRAQLEEMARVAAEPMARARDDDAMNAHLKEVLHAADPMA--------NM 386
Query 114 GRKQKGKEDVDREKKHQRPKCPHDAPPNRFGIKPGYRWDGVVRGNGYE 161
+K++ +DR + P PNRFGI PGYRWDGV R NG+E
Sbjct 387 IQKKRRDTAIDR-GELVYPSYHGHFVPNRFGIAPGYRWDGVDRSNGFE 433
> YGL174w
Length=266
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 18/25 (72%), Positives = 19/25 (76%), Gaps = 0/25 (0%)
Query 138 APPNRFGIKPGYRWDGVVRGNGYEE 162
AP NRF I PG RWDGV R NG+EE
Sbjct 219 APENRFAIMPGSRWDGVHRSNGFEE 243
> YHR154w
Length=1070
Score = 31.6 bits (70), Expect = 0.84, Method: Composition-based stats.
Identities = 17/69 (24%), Positives = 32/69 (46%), Gaps = 0/69 (0%)
Query 65 EARKKEEEKIAKQPFARYDIDEEHDAELRARDRWDDPLAKAPSNTAQGLGRKQKGKEDVD 124
E +K EEK +P YD ++ + ++R + D K T + Q+ K ++
Sbjct 604 EGHEKREEKEFTKPITEYDAPKKQEIREQSRKKNDIDYKKEEEETELQVQLGQRTKREIK 663
Query 125 REKKHQRPK 133
KK+++ K
Sbjct 664 TSKKNEKEK 672
> 7297351
Length=2771
Score = 29.3 bits (64), Expect = 4.0, Method: Composition-based stats.
Identities = 27/96 (28%), Positives = 41/96 (42%), Gaps = 11/96 (11%)
Query 43 QRERGPAPVLEWGAGLKQKEDLEARKKEEEKIAKQPFARYDIDEEHDAELRA-------- 94
+R + VL +GL + L +K ++ + P I +E DA +
Sbjct 1587 RRSQTLLEVLLPASGLTRYIYLNPQKSKDAPTGEFPSTSKGISKERDAATASSSASPAPT 1646
Query 95 RDRWDDPLAKAPSNTAQGLGRKQKGKEDVDREKKHQ 130
RD D P+ PS GLGR+Q KE D + K +
Sbjct 1647 RDVGDLPVIPPPST---GLGREQTSKETSDSKSKME 1679
Lambda K H
0.313 0.135 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2562785186
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40