bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2288_orf2
Length=149
Score E
Sequences producing significant alignments: (Bits) Value
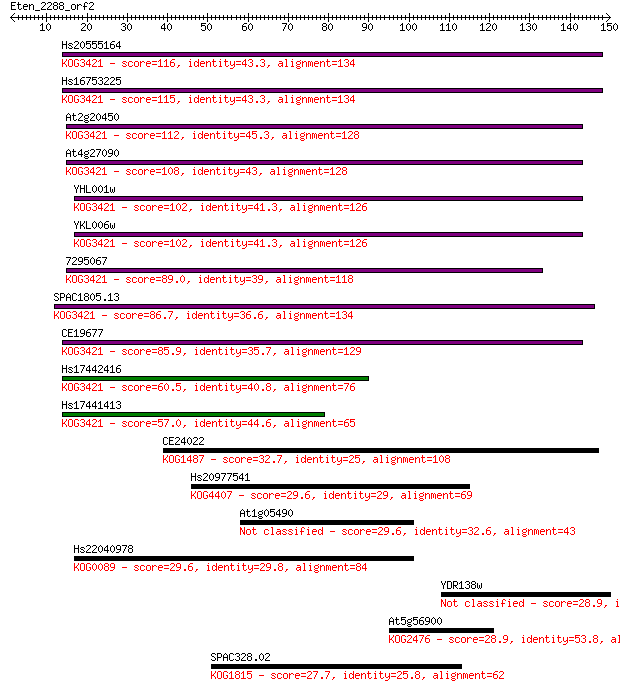
Hs20555164 116 1e-26
Hs16753225 115 2e-26
At2g20450 112 2e-25
At4g27090 108 3e-24
YHL001w 102 4e-22
YKL006w 102 4e-22
7295067 89.0 3e-18
SPAC1805.13 86.7 2e-17
CE19677 85.9 3e-17
Hs17442416 60.5 1e-09
Hs17441413 57.0 1e-08
CE24022 32.7 0.30
Hs20977541 29.6 2.2
At1g05490 29.6 2.4
Hs22040978 29.6 2.4
YDR138w 28.9 3.8
At5g56900 28.9 4.0
SPAC328.02 27.7 8.8
> Hs20555164
Length=215
Score = 116 bits (291), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 58/134 (43%), Positives = 91/134 (67%), Gaps = 2/134 (1%)
Query 14 LFRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALTDM 73
+FRRF+E GR+ V +GP GKL +VD+I+ NR LVDG T VRRQ++P + + LTD
Sbjct 2 VFRRFVEVGRVAYVSFGPHAGKLVAIVDVIDQNRALVDGP-CTQVRRQAMPFKCMQLTDF 60
Query 74 RLKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQRA 133
LK PH R +++A +K D+ K+ + W +K A++R+A +TDF+RFK+M A+K R
Sbjct 61 ILKFPHSARQKYVQQAWQKADINTKWAATRWAKKIEARERKAKMTDFDRFKVMKAKKTRN 120
Query 134 QAVRAKLGKRMQKV 147
+ ++ ++ K++QK
Sbjct 121 RIIKNEV-KKLQKA 133
> Hs16753225
Length=216
Score = 115 bits (289), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 58/134 (43%), Positives = 90/134 (67%), Gaps = 2/134 (1%)
Query 14 LFRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALTDM 73
+FRRF+E GR+ V +GP GKL +VD+I+ NR LVDG T VRRQ++P + + LTD
Sbjct 2 VFRRFVEVGRVAYVSFGPHAGKLVAIVDVIDQNRALVDGP-CTQVRRQAMPFKCMQLTDF 60
Query 74 RLKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQRA 133
LK PH +R+A +K D+ K+ + W +K A++R+A +TDF+RFK+M A+K R
Sbjct 61 ILKFPHSAHQKYVRQAWQKADINTKWAATRWAKKIEARERKAKMTDFDRFKVMKAKKMRN 120
Query 134 QAVRAKLGKRMQKV 147
+ ++ ++ K++QK
Sbjct 121 RIIKNEV-KKLQKA 133
> At2g20450
Length=134
Score = 112 bits (281), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 58/128 (45%), Positives = 88/128 (68%), Gaps = 3/128 (2%)
Query 15 FRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALTDMR 74
F+RF+E GR+ +V YG D GKL +VD+++ NR LVD ++ R + ++R++LTD+
Sbjct 3 FKRFVEIGRVALVNYGEDYGKLVVIVDVVDQNRALVDAPDME---RIQMNLKRLSLTDIV 59
Query 75 LKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQRAQ 134
+ I + L +A+EK DV K+ +SSWGRK I ++RRAAL DF+RFK+M+A+ +RA
Sbjct 60 IDINRVPKKKVLIEAMEKADVKNKWEKSSWGRKLIVQKRRAALNDFDRFKIMLAKIKRAG 119
Query 135 AVRAKLGK 142
VR +L K
Sbjct 120 IVRQELAK 127
> At4g27090
Length=134
Score = 108 bits (271), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 55/128 (42%), Positives = 86/128 (67%), Gaps = 3/128 (2%)
Query 15 FRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALTDMR 74
F+R++E GR+ +V YG D GKL +VD+++ NR LVD ++ R + +R++LTD+
Sbjct 3 FKRYVEIGRVALVNYGEDHGKLVVIVDVVDQNRALVDAPDM---ERIQMNFKRLSLTDIV 59
Query 75 LKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQRAQ 134
+ I + L +A+EK DV K+ +SSWGRK I ++RRA L DF+RFK+M+A+ ++A
Sbjct 60 IDINRVPKKKALIEAMEKADVKNKWEKSSWGRKLIVQKRRANLNDFDRFKIMLAKIKKAG 119
Query 135 AVRAKLGK 142
VR +L K
Sbjct 120 VVRQELAK 127
> YHL001w
Length=138
Score = 102 bits (253), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 52/126 (41%), Positives = 79/126 (62%), Gaps = 1/126 (0%)
Query 17 RFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALTDMRLK 76
R +E GR+ +++ G GKL +V+II+ ++L+DG GV RQ+I + +V LT +
Sbjct 13 RLVEVGRVVLIKKGQSAGKLAAIVEIIDQKKVLIDGPK-AGVPRQAINLGQVVLTPLTFA 71
Query 77 IPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQRAQAV 136
+P G R+ T+ K V +K+ SSW +K ++RRAALTDFERF++MV RKQ+ V
Sbjct 72 LPRGARTATVSKKWAAAGVCEKWAASSWAKKIAQRERRAALTDFERFQVMVLRKQKRYTV 131
Query 137 RAKLGK 142
+ L K
Sbjct 132 KKALAK 137
> YKL006w
Length=138
Score = 102 bits (253), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 52/126 (41%), Positives = 79/126 (62%), Gaps = 1/126 (0%)
Query 17 RFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALTDMRLK 76
R +E GR+ +++ G GKL +V+II+ ++L+DG GV RQ+I + +V LT +
Sbjct 13 RLVEVGRVVLIKKGQSAGKLAAIVEIIDQKKVLIDGPK-AGVPRQAINLGQVVLTPLTFA 71
Query 77 IPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQRAQAV 136
+P G R+ T+ K V +K+ SSW +K ++RRAALTDFERF++MV RKQ+ V
Sbjct 72 LPRGARTATVSKKWAAAAVCEKWAASSWAKKIAQRERRAALTDFERFQVMVLRKQKRYTV 131
Query 137 RAKLGK 142
+ L K
Sbjct 132 KKALAK 137
> 7295067
Length=166
Score = 89.0 bits (219), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 46/118 (38%), Positives = 72/118 (61%), Gaps = 1/118 (0%)
Query 15 FRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALTDMR 74
F RF++ GR+ GP +G+L +VD+I+ NR+LVDG +TGV RQ + + LT R
Sbjct 3 FERFVQTGRIAKASAGPLKGRLVAIVDVIDQNRVLVDGP-LTGVPRQEYRLNNLHLTKYR 61
Query 75 LKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQR 132
+K P+ + +RKA + D+ ++ S W K +R++L DF+RFKL A++QR
Sbjct 62 IKFPYTAPTRIVRKAWTESDLKAQWKVSPWSVKAQNICKRSSLNDFDRFKLRYAKRQR 119
> SPAC1805.13
Length=134
Score = 86.7 bits (213), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 49/134 (36%), Positives = 77/134 (57%), Gaps = 1/134 (0%)
Query 12 MALFRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALT 71
M F+R++E GR+ +V G GKL +VDI++ R L+D + + RQ I V LT
Sbjct 1 MEGFKRYVEVGRVVLVTKGEYTGKLAVIVDIVDHKRALID-SPCSEFPRQVIRYGSVVLT 59
Query 72 DMRLKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQ 131
+ +K+P G RS + K + DV K+ S+W +K AK+ R+ L DF+RF +M +KQ
Sbjct 60 HIVMKLPRGARSGIVAKKWKAQDVCNKWASSAWAKKLEAKKVRSQLNDFDRFAVMRLKKQ 119
Query 132 RAQAVRAKLGKRMQ 145
R + V + K ++
Sbjct 120 RREQVNVAVAKALK 133
> CE19677
Length=135
Score = 85.9 bits (211), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 46/129 (35%), Positives = 78/129 (60%), Gaps = 1/129 (0%)
Query 14 LFRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALTDM 73
+F R ++ GR+ + G D+GKL +V++I+GNR+ +DG + + V R ++ + LT
Sbjct 2 VFNRVVQIGRVVFIASGKDQGKLAAIVNVIDGNRVQIDGPS-SDVTRTVRNLKDLQLTKF 60
Query 74 RLKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQRA 133
LK+ G R+ ++ A + V + F ++ W +K + RA LTDFER+KLM A++ R
Sbjct 61 VLKLRVGQRTKGVKAAFDAAKVTENFQKTQWAKKIAQRAIRAKLTDFERYKLMKAKQMRN 120
Query 134 QAVRAKLGK 142
+ VR +L K
Sbjct 121 RIVRVELAK 129
> Hs17442416
Length=145
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 31/76 (40%), Positives = 43/76 (56%), Gaps = 1/76 (1%)
Query 14 LFRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALTDM 73
+F+ F+E ++ V +GP GKL VVD+ + NR L DG T V Q++ + + LTD
Sbjct 2 MFKHFLEVCQVAYVSFGPYAGKLVAVVDVTDENRALADGL-CTQVMGQAVSFKCMQLTDF 60
Query 74 RLKIPHGVRSVTLRKA 89
LK PH LRKA
Sbjct 61 ILKFPHSTHQRKLRKA 76
> Hs17441413
Length=134
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 29/65 (44%), Positives = 43/65 (66%), Gaps = 2/65 (3%)
Query 14 LFRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALTDM 73
+FR F+E G++ + GP GKL ++D+++ NR LV GA T VRRQ++P + + LTD
Sbjct 2 VFRHFVEVGQVAYIS-GPHAGKLVAIIDVVDENRALVGGA-CTQVRRQAMPFKCMQLTDF 59
Query 74 RLKIP 78
LK P
Sbjct 60 ILKFP 64
> CE24022
Length=366
Score = 32.7 bits (73), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 27/108 (25%), Positives = 52/108 (48%), Gaps = 6/108 (5%)
Query 39 VVDIINGNRILVDGANVTGVRRQSIPIRRVALTDMRLKIPHGVRSVTLRKALEKDDVIKK 98
++D+I GNRI + V + I I + D+ +IPH V ++ DD+++K
Sbjct 228 LIDVIEGNRIYIPCIYVLN-KIDQISIEEL---DIIYRIPHTV-PISAHHKWNFDDLLEK 282
Query 99 FNESSWGRKRIAKQRRAALTDFERFKLMVARKQRAQAVRAKLGKRMQK 146
E RI + + L D+ + ++ A ++ + + K+ K +QK
Sbjct 283 VWE-YLNLIRIYTKPKGQLPDYSQPIVLNAERKSIEDLCTKIHKSLQK 329
> Hs20977541
Length=1957
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 20/72 (27%), Positives = 34/72 (47%), Gaps = 4/72 (5%)
Query 46 NRILVDGANVTGVRRQSIPIRRVAL---TDMRLKIPHGVRSVTLRKALEKDDVIKKFNES 102
N ++ + RQS+ IR+ L ++ + + PH + + RK L KDD ++
Sbjct 1061 NNLMSKAEQLPKTPRQSLSIRQTLLGAKSEPKTQSPHSPKEESERKLLSKDDTSPPKDKG 1120
Query 103 SWGRKRIAKQRR 114
+W RK I R
Sbjct 1121 TW-RKGIPSIMR 1131
> At1g05490
Length=731
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 26/43 (60%), Gaps = 0/43 (0%)
Query 58 VRRQSIPIRRVALTDMRLKIPHGVRSVTLRKALEKDDVIKKFN 100
+R+ S PI+RV+L + + + + +L K E+D+ I+K N
Sbjct 321 LRKASSPIKRVSLVERKALVRYKRSGSSLTKPRERDNKIQKLN 363
> Hs22040978
Length=402
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 25/87 (28%), Positives = 35/87 (40%), Gaps = 3/87 (3%)
Query 17 RFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQ---SIPIRRVALTDM 73
I GR+C+ QY C V +II I G NV R +PI + TD
Sbjct 239 HVINAGRMCLDQYSMLPATPCGVWEIIKRTDIPTLGKNVVVAGRSKSVEMPIAMLLHTDG 298
Query 74 RLKIPHGVRSVTLRKALEKDDVIKKFN 100
+ P G +VT+ + +KK
Sbjct 299 VHERPGGDATVTISYRYTSKEQLKKHT 325
> YDR138w
Length=752
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 23/42 (54%), Gaps = 0/42 (0%)
Query 108 RIAKQRRAALTDFERFKLMVARKQRAQAVRAKLGKRMQKVES 149
+I ++ R L D +K+ RK+RA A +R QK++S
Sbjct 627 KINQEHRKKLQDAREYKIGKERKKRALEEEASFPEREQKIKS 668
> At5g56900
Length=593
Score = 28.9 bits (63), Expect = 4.0, Method: Composition-based stats.
Identities = 14/26 (53%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 95 VIKKFNESSWGRKRIAKQRRAALTDF 120
V KKFN+S+ GRK + K+ AAL F
Sbjct 499 VTKKFNDSTDGRKYLQKEYNAALGLF 524
> SPAC328.02
Length=504
Score = 27.7 bits (60), Expect = 8.8, Method: Composition-based stats.
Identities = 16/64 (25%), Positives = 32/64 (50%), Gaps = 2/64 (3%)
Query 51 DGANVTGVRRQ--SIPIRRVALTDMRLKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKR 108
DG V RR+ S+ R V++ D+R + + +T L ++ V+ + W R+R
Sbjct 37 DGFTVERKRRRAHSVSYRVVSVRDLRASLNEKINQLTSIIDLTREQVLGLYRYFKWNRER 96
Query 109 IAKQ 112
+ ++
Sbjct 97 LLER 100
Lambda K H
0.325 0.139 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1858150626
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40