bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
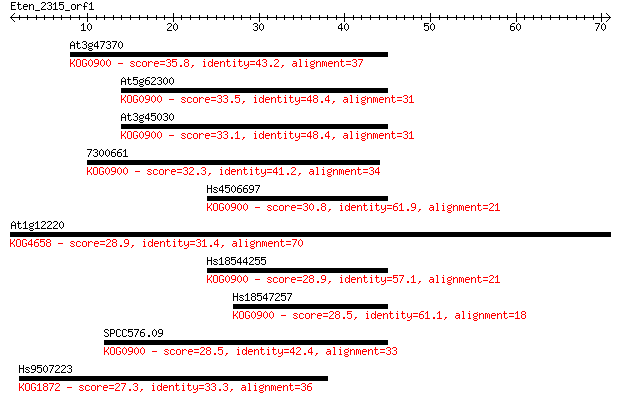
Query= Eten_2315_orf1
Length=70
Score E
Sequences producing significant alignments: (Bits) Value
At3g47370 35.8 0.019
At5g62300 33.5 0.11
At3g45030 33.1 0.14
7300661 32.3 0.23
Hs4506697 30.8 0.58
At1g12220 28.9 2.2
Hs18544255 28.9 2.4
Hs18547257 28.5 3.1
SPCC576.09 28.5 3.1
Hs9507223 27.3 6.8
> At3g47370
Length=122
Score = 35.8 bits (81), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 27/37 (72%), Gaps = 0/37 (0%)
Query 8 LAASMSKALKEGLEGEEQRLHRIRITLTSKDLKSIER 44
+A K K GLE +++H+IRITL+SK++K++E+
Sbjct 1 MAYEPMKPTKAGLEAPLEQIHKIRITLSSKNVKNLEK 37
> At5g62300
Length=124
Score = 33.5 bits (75), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 15/31 (48%), Positives = 25/31 (80%), Gaps = 0/31 (0%)
Query 14 KALKEGLEGEEQRLHRIRITLTSKDLKSIER 44
K K GLE +++H+IRITL+SK++K++E+
Sbjct 9 KPGKAGLEEPLEQIHKIRITLSSKNVKNLEK 39
> At3g45030
Length=117
Score = 33.1 bits (74), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 15/31 (48%), Positives = 25/31 (80%), Gaps = 0/31 (0%)
Query 14 KALKEGLEGEEQRLHRIRITLTSKDLKSIER 44
K K GLE +++H+IRITL+SK++K++E+
Sbjct 2 KPGKAGLEEPLEQIHKIRITLSSKNVKNLEK 32
> 7300661
Length=120
Score = 32.3 bits (72), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 25/34 (73%), Gaps = 0/34 (0%)
Query 10 ASMSKALKEGLEGEEQRLHRIRITLTSKDLKSIE 43
A+ K +++ G+ +HRIRITLTS++++S+E
Sbjct 2 AAAPKDIEKPHVGDSASVHRIRITLTSRNVRSLE 35
> Hs4506697
Length=119
Score = 30.8 bits (68), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 13/21 (61%), Positives = 19/21 (90%), Gaps = 0/21 (0%)
Query 24 EQRLHRIRITLTSKDLKSIER 44
E +HRIRITLTS+++KS+E+
Sbjct 14 EVAIHRIRITLTSRNVKSLEK 34
> At1g12220
Length=889
Score = 28.9 bits (63), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 22/85 (25%), Positives = 38/85 (44%), Gaps = 15/85 (17%)
Query 1 FIHPFSSLAASMSKALK---------------EGLEGEEQRLHRIRITLTSKDLKSIERG 45
+IH S AS+ KA++ E G +QRL ++++ LTS + +
Sbjct 28 YIHNLSKNLASLQKAMRMLKARQYDVIRRLETEEFTGRQQRLSQVQVWLTSVLIIQNQFN 87
Query 46 EQQRKNDISAFLEVLKPFCSRYMGL 70
+ R N++ L FCS+ + L
Sbjct 88 DLLRSNEVELQRLCLCGFCSKDLKL 112
> Hs18544255
Length=130
Score = 28.9 bits (63), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 12/21 (57%), Positives = 19/21 (90%), Gaps = 0/21 (0%)
Query 24 EQRLHRIRITLTSKDLKSIER 44
E +H+IRITLTS+++KS+E+
Sbjct 25 EVAIHQIRITLTSRNVKSLEK 45
> Hs18547257
Length=149
Score = 28.5 bits (62), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 11/18 (61%), Positives = 17/18 (94%), Gaps = 0/18 (0%)
Query 27 LHRIRITLTSKDLKSIER 44
+HRIRIT+TS+ +KS+E+
Sbjct 48 IHRIRITVTSRKVKSLEK 65
> SPCC576.09
Length=118
Score = 28.5 bits (62), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 25/34 (73%), Gaps = 1/34 (2%)
Query 12 MSKALKEGLEGE-EQRLHRIRITLTSKDLKSIER 44
MS+ K+ E + +HRIRITLTS++++++E+
Sbjct 1 MSQVAKDQKEQQIPSTVHRIRITLTSRNVRNLEK 34
> Hs9507223
Length=533
Score = 27.3 bits (59), Expect = 6.8, Method: Composition-based stats.
Identities = 12/36 (33%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 2 IHPFSSLAASMSKALKEGLEGEEQRLHRIRITLTSK 37
IH + L +M K + +GL E++ L I++T +K
Sbjct 338 IHSITGLPPAMQKVMYKGLVPEDKTLREIKVTSGAK 373
Lambda K H
0.320 0.134 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1197219688
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40