bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
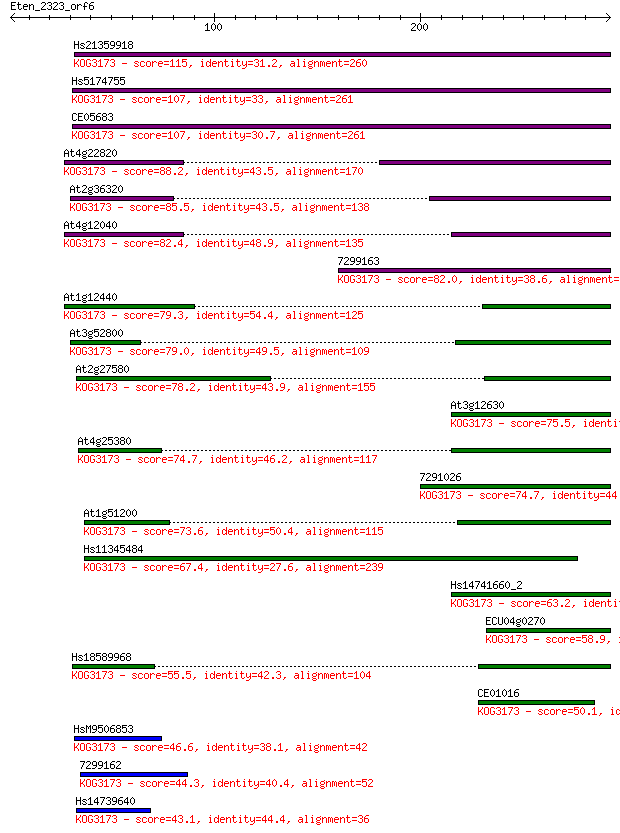
Query= Eten_2323_orf6
Length=291
Score E
Sequences producing significant alignments: (Bits) Value
Hs21359918 115 1e-25
Hs5174755 107 2e-23
CE05683 107 2e-23
At4g22820 88.2 2e-17
At2g36320 85.5 1e-16
At4g12040 82.4 9e-16
7299163 82.0 1e-15
At1g12440 79.3 8e-15
At3g52800 79.0 1e-14
At2g27580 78.2 2e-14
At3g12630 75.5 1e-13
At4g25380 74.7 2e-13
7291026 74.7 2e-13
At1g51200 73.6 5e-13
Hs11345484 67.4 4e-11
Hs14741660_2 63.2 6e-10
ECU04g0270 58.9 1e-08
Hs18589968 55.5 1e-07
CE01016 50.1 6e-06
HsM9506853 46.6 6e-05
7299162 44.3 3e-04
Hs14739640 43.1 7e-04
> Hs21359918
Length=208
Score = 115 bits (288), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 81/267 (30%), Positives = 122/267 (45%), Gaps = 68/267 (25%)
Query 32 HENERPSAPPLCAKNCGFYGSPANRNLCSKCYREFLKAESAA-------ATAAAACRRAL 84
E P LC+ CGFYG+P +CS CY+E L+ ++++ AT+ ++ +L
Sbjct 3 QETNHSQVPMLCSTGCGFYGNPRTNGMCSVCYKEHLQRQNSSNGRISPPATSVSSLSESL 62
Query 85 PQQQQNSDAVAGDTSSAVPAEQSSAARSQLKAVTEPSDAIGPAAVAATEATAAAATVAAE 144
P Q T +VP QS A+ +T ++ + V+ +
Sbjct 63 PVQ---------CTDGSVPEAQS--------------------ALDSTSSSMQPSPVSNQ 93
Query 145 SCGEEATAPTQEPADDAAAVGLAPAAVPSTASDPQPGVAATPEATAAAAAATSMTSSSPV 204
S E+ A +Q + ST+ D A PE A+ +
Sbjct 94 SLLSESVASSQ---------------LDSTSVD-----KAVPETEDVQASVS-------- 125
Query 205 ESTDTAAPPSPSSAPASSSQGRGKTSRCHKCNKKVGLLGFVCRCGFAFCGEHRYADAHGC 264
DTA PS + S + + K +RC C KKVGL GF CRCG +CG HRY+D H C
Sbjct 126 ---DTAQQPSEEQS-KSLEKPKQKKNRCFMCRKKVGLTGFECRCGNVYCGVHRYSDVHNC 181
Query 265 VFDYKAFEREQLRKQNNKVVADKLQKL 291
++YKA E++RK+N VV +K+QK+
Sbjct 182 SYNYKADAAEKIRKENPVVVGEKIQKI 208
> Hs5174755
Length=213
Score = 107 bits (268), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 86/269 (31%), Positives = 120/269 (44%), Gaps = 66/269 (24%)
Query 31 QHENERPSAPPLCAKNCGFYGSPANRNLCSKCYREFLKAESAAATAAAACRRALPQQQQN 90
Q N+ P P LC+ CGFYG+P +CS CY+E L Q+QQN
Sbjct 3 QETNQTP-GPMLCSTGCGFYGNPRTNGMCSVCYKEHL------------------QRQQN 43
Query 91 SDAVAGDTSSAVPAEQSSAARSQLKAVTEPSDAIGPAAVAATEATAAAATVAAESCGEEA 150
S ++ P +S + S SD+ A+ A + +C E A
Sbjct 44 SGRMS-------PMGTASGSNSPT------SDS----------ASVQRADTSLNNC-EGA 79
Query 151 TAPTQEPADDAAAVGLAPAAVPSTASDPQPGVAATPEATAAAAAATSMTSSSPVESTDTA 210
T E + + AA+P T + ++ + T S PV
Sbjct 80 AGSTSEKSRNVPV-----AALPVTQQMTEMSISREDKITTPKTEV-----SEPV-----V 124
Query 211 APPSPSSAPASSSQG--------RGKTSRCHKCNKKVGLLGFVCRCGFAFCGEHRYADAH 262
PSPS + S+SQ + K +RC C KKVGL GF CRCG FCG HRY+D H
Sbjct 125 TQPSPSVSQPSTSQSEEKAPELPKPKKNRCFMCRKKVGLTGFDCRCGNLFCGLHRYSDKH 184
Query 263 GCVFDYKAFEREQLRKQNNKVVADKLQKL 291
C +DYKA ++RK+N VVA+K+Q++
Sbjct 185 NCPYDYKAEAAAKIRKENPVVVAEKIQRI 213
> CE05683
Length=189
Score = 107 bits (268), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 80/263 (30%), Positives = 113/263 (42%), Gaps = 77/263 (29%)
Query 31 QHENERPSAPPLCAKNCGFYGSPANRNLCSKCYREFLKAESAAATAAAACRRALPQQQQN 90
++E ++ P C CGF+G+ A CS+C++ LK +QQ+
Sbjct 2 ENEQQQAQTAPSCRAGCGFFGASATEGYCSQCFKNTLK------------------RQQD 43
Query 91 SDAVAGDTSSAVPAEQSSAARSQLKAVTEPS--DAIGPAAVAATEATAAAATVAAESCGE 148
+ + TS V +A S LK+ EPS D AAV+ ++ TA E
Sbjct 44 TVRL---TSPVVSPSSMAATSSALKS--EPSSVDMCMKAAVSVSDETAKMDC-------E 91
Query 149 EATAPTQEPADDAAAVGLAPAAVPSTASDPQPGVAATPEATAAAAAATSMTSSSPVESTD 208
+ + DD+ V A + A T++T PV
Sbjct 92 DIINVCDQINDDSVTV-------------------------AESTAPTTITVDVPV---- 122
Query 209 TAAPPSPSSAPASSSQGRGKTSRCHKCNKKVGLLGFVCRCGFAFCGEHRYADAHGCVFDY 268
P K +RCH C K+VGL GF CRCG +CG+HRY AH C FDY
Sbjct 123 ----------PVK------KANRCHMCKKRVGLTGFSCRCGGLYCGDHRYDQAHNCQFDY 166
Query 269 KAFEREQLRKQNNKVVADKLQKL 291
K ERE +RK N VV+DK+Q++
Sbjct 167 KTMERETIRKNNPVVVSDKVQRI 189
> At4g22820
Length=176
Score = 88.2 bits (217), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 46/113 (40%), Positives = 61/113 (53%), Gaps = 14/113 (12%)
Query 180 PGVAATPEATAAAAAATSMTSSSPVESTDTAAPPSPSSAPASSSQGR-GKTSRCHKCNKK 238
P V P+ AA S A PS S+ P ++ R +T+RC CNKK
Sbjct 77 PAVVVEPKPEKAAVVVVS-------------AEPSSSAVPEANEPSRPARTNRCLCCNKK 123
Query 239 VGLLGFVCRCGFAFCGEHRYADAHGCVFDYKAFEREQLRKQNNKVVADKLQKL 291
VG++GF C+CG FCGEHRY + H C FD+K R ++ K N V ADK+Q+
Sbjct 124 VGIMGFKCKCGSTFCGEHRYPETHDCSFDFKEVGRGEIAKANPVVKADKIQRF 176
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 28/63 (44%), Positives = 38/63 (60%), Gaps = 5/63 (7%)
Query 27 MSSEQHEN-----ERPSAPPLCAKNCGFYGSPANRNLCSKCYREFLKAESAAATAAAACR 81
M SEQ+++ + S P LC K CGF+GSP+N +LCSKCYR E+ A A AA
Sbjct 1 MGSEQNDSTSFTQSQASEPKLCVKGCGFFGSPSNMDLCSKCYRGICAEEAQTAVAKAAVE 60
Query 82 RAL 84
++
Sbjct 61 KSF 63
> At2g36320
Length=161
Score = 85.5 bits (210), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 38/88 (43%), Positives = 51/88 (57%), Gaps = 0/88 (0%)
Query 204 VESTDTAAPPSPSSAPASSSQGRGKTSRCHKCNKKVGLLGFVCRCGFAFCGEHRYADAHG 263
+ +T P P + +RC C K+VGL GF+CRCG FCG HRY + HG
Sbjct 74 IPTTKKTEEKKPIQIPTEQPSPPQRPNRCTVCRKRVGLTGFMCRCGTTFCGSHRYPEVHG 133
Query 264 CVFDYKAFEREQLRKQNNKVVADKLQKL 291
C FD+K+ RE++ K N V+A KLQK+
Sbjct 134 CTFDFKSAGREEIAKANPLVIAAKLQKI 161
Score = 47.0 bits (110), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 22/51 (43%), Positives = 28/51 (54%), Gaps = 1/51 (1%)
Query 30 EQHENERPSAPPLCAKNCGFYGSPANRNLCSKCYREF-LKAESAAATAAAA 79
E+H E P LC NCGF+GS A NLCS CY + LK + A+ +
Sbjct 3 EEHRCETPEGHRLCVNNCGFFGSSATMNLCSNCYGDLCLKQQQQASMKSTV 53
> At4g12040
Length=175
Score = 82.4 bits (202), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 38/84 (45%), Positives = 51/84 (60%), Gaps = 7/84 (8%)
Query 215 PSSAPASSSQGRGKTSR-------CHKCNKKVGLLGFVCRCGFAFCGEHRYADAHGCVFD 267
PSS P ++ Q + SR C CNKKVG++GF C+CG FCG HRY + H C FD
Sbjct 92 PSSVPVAAEQDEAEPSRPVRPNNRCFSCNKKVGVMGFKCKCGSTFCGSHRYPEKHECSFD 151
Query 268 YKAFEREQLRKQNNKVVADKLQKL 291
+K R+ + K N V ADK+Q++
Sbjct 152 FKEVGRDAIAKANPLVKADKVQRI 175
Score = 57.0 bits (136), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 28/60 (46%), Positives = 36/60 (60%), Gaps = 2/60 (3%)
Query 27 MSSEQHENER--PSAPPLCAKNCGFYGSPANRNLCSKCYREFLKAESAAATAAAACRRAL 84
M SE++ + P+ P LC CGF+GSP+N NLCSKCYR E A A AA + +L
Sbjct 1 MGSEENNSTSFPPTEPKLCDNGCGFFGSPSNMNLCSKCYRSLRAEEDQTAVAKAAVKNSL 60
> 7299163
Length=178
Score = 82.0 bits (201), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 51/144 (35%), Positives = 73/144 (50%), Gaps = 19/144 (13%)
Query 160 DAAAVGLAPAAVPSTASDPQPGVAATPEATAAAAAATSMTSSSPVESTDTAAPPSPSSAP 219
D +V +A V ASD V + T AAAA + S E+ +A P+ S+
Sbjct 42 DVGSVKMADV-VLEGASDV--AVTKKRKITEEAAAAAKVNS----EAITSATGPNTSTQA 94
Query 220 ASSSQGRGKT------------SRCHKCNKKVGLLGFVCRCGFAFCGEHRYADAHGCVFD 267
A+S+ +RC +C KKVGL GF CRCG +C HRY+D H C FD
Sbjct 95 AASANEEDDKDKEDDKDAKKKKNRCGECRKKVGLTGFQCRCGGLYCAVHRYSDKHNCTFD 154
Query 268 YKAFEREQLRKQNNKVVADKLQKL 291
Y+ +++R+ N VV +K+QK+
Sbjct 155 YREHGAQEIRRNNPVVVGEKIQKI 178
> At1g12440
Length=168
Score = 79.3 bits (194), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 33/62 (53%), Positives = 42/62 (67%), Gaps = 0/62 (0%)
Query 230 SRCHKCNKKVGLLGFVCRCGFAFCGEHRYADAHGCVFDYKAFEREQLRKQNNKVVADKLQ 289
+RC CNKKVG+ GF CRCG FCG HRY ++H C FD+K RE + K N V ADK+
Sbjct 107 TRCLSCNKKVGVTGFKCRCGSTFCGTHRYPESHECQFDFKGVAREAIAKANPVVKADKVD 166
Query 290 KL 291
++
Sbjct 167 RI 168
Score = 65.9 bits (159), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 35/73 (47%), Positives = 44/73 (60%), Gaps = 10/73 (13%)
Query 27 MSSEQHENER--PSAPPLCAKNCGFYGSPANRNLCSKCYREFLKAESAAATAAAACRRAL 84
M SEQ+++ PS P LC K CGF+GSP+N NLCSKCYR+ E A+A AA ++L
Sbjct 1 MGSEQNDSTSFSPSEPKLCVKGCGFFGSPSNMNLCSKCYRDIRATEEQTASAKAAVEKSL 60
Query 85 --------PQQQQ 89
PQQ Q
Sbjct 61 NPNKPKTQPQQSQ 73
> At3g52800
Length=170
Score = 79.0 bits (193), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 36/75 (48%), Positives = 48/75 (64%), Gaps = 0/75 (0%)
Query 217 SAPASSSQGRGKTSRCHKCNKKVGLLGFVCRCGFAFCGEHRYADAHGCVFDYKAFEREQL 276
S P + + + +RC C K+VGL GF CRCG FCG HRY + HGC +D+K+ RE++
Sbjct 96 SLPTTEQNQQQRPNRCTTCRKRVGLTGFKCRCGTMFCGVHRYPEIHGCSYDFKSAGREEI 155
Query 277 RKQNNKVVADKLQKL 291
K N V A KLQK+
Sbjct 156 AKANPLVKAAKLQKI 170
Score = 43.9 bits (102), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 18/34 (52%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 30 EQHENERPSAPPLCAKNCGFYGSPANRNLCSKCY 63
E+H + P + LC NCGF GS A NLCS CY
Sbjct 3 EEHRCQTPESNRLCVNNCGFLGSSATMNLCSNCY 36
> At2g27580
Length=163
Score = 78.2 bits (191), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 32/61 (52%), Positives = 43/61 (70%), Gaps = 0/61 (0%)
Query 231 RCHKCNKKVGLLGFVCRCGFAFCGEHRYADAHGCVFDYKAFEREQLRKQNNKVVADKLQK 290
RC C ++VG+ GF CRCGF FCG HRYA+ H C FD+K ++++ K N V ADKL+K
Sbjct 103 RCLTCRRRVGITGFRCRCGFVFCGTHRYAEQHECSFDFKRMGKDKIAKANPIVKADKLEK 162
Query 291 L 291
+
Sbjct 163 I 163
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 36/99 (36%), Positives = 52/99 (52%), Gaps = 9/99 (9%)
Query 33 ENERPSAPPLCAKNCGFYGSPANRNLCSKCYREFLKAESAAATAAAACRRALPQ-----Q 87
E R P LCA NCGF+GS A +NLCSKC+R+ E ++TA + AL Q
Sbjct 3 EEHRLQEPRLCANNCGFFGSTATQNLCSKCFRDLQHQEQNSSTA----KHALTQSLAAVG 58
Query 88 QQNSDAVAGDTSSAVPAEQSSAARSQLKAVTEPSDAIGP 126
S +V+ +++ A+S+ +A EP +A GP
Sbjct 59 AAASSSVSPPPPPPADSKEIVEAKSEKRAAAEPEEADGP 97
> At3g12630
Length=160
Score = 75.5 bits (184), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 35/77 (45%), Positives = 45/77 (58%), Gaps = 0/77 (0%)
Query 215 PSSAPASSSQGRGKTSRCHKCNKKVGLLGFVCRCGFAFCGEHRYADAHGCVFDYKAFERE 274
P A + + +RC C KKVGL GF CRCG FC EHRY+D H C +DYK RE
Sbjct 84 PREIDAVKKRDQQIVNRCSGCRKKVGLTGFRCRCGELFCSEHRYSDRHDCSYDYKTAGRE 143
Query 275 QLRKQNNKVVADKLQKL 291
+ ++N V A K+ K+
Sbjct 144 AIARENPVVKAAKMVKV 160
> At4g25380
Length=130
Score = 74.7 bits (182), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 37/77 (48%), Positives = 46/77 (59%), Gaps = 1/77 (1%)
Query 215 PSSAPASSSQGRGKTSRCHKCNKKVGLLGFVCRCGFAFCGEHRYADAHGCVFDYKAFERE 274
P+++PA + K RC C +KVG+LGF CRCG FCG HRY + H C FDYK R
Sbjct 55 PTNSPAVEEEP-VKKRRCGICKRKVGMLGFKCRCGHMFCGSHRYPEEHSCPFDYKQSGRL 113
Query 275 QLRKQNNKVVADKLQKL 291
L Q + ADKLQ+
Sbjct 114 ALATQLPLIRADKLQRF 130
Score = 33.1 bits (74), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 17/40 (42%), Positives = 22/40 (55%), Gaps = 2/40 (5%)
Query 34 NERPSAPPLCAKNCGFYGSPANRNLCSKCYREFLKAESAA 73
NE + P C CG YG+ N NLCS CY++ + S A
Sbjct 3 NETEALP--CEGGCGLYGTRVNNNLCSLCYKKSVLQHSPA 40
> 7291026
Length=162
Score = 74.7 bits (182), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 41/97 (42%), Positives = 50/97 (51%), Gaps = 5/97 (5%)
Query 200 SSSPVESTDTAAPPSPSSAPASSSQGR----GKTSRCHKCNKKVGLLG-FVCRCGFAFCG 254
S V S D AA S+ G RC KC KK+G+ G F CRCG +C
Sbjct 66 DSRAVNSKDGAANQDTSADSGQDQDGNQAQDSTKKRCDKCGKKLGITGGFPCRCGGTYCA 125
Query 255 EHRYADAHGCVFDYKAFEREQLRKQNNKVVADKLQKL 291
HRY+D H C FDY+ Q+R+ N VVA KL+KL
Sbjct 126 VHRYSDRHECNFDYRQMGAIQIRRDNPVVVASKLRKL 162
> At1g51200
Length=173
Score = 73.6 bits (179), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 39/74 (52%), Positives = 49/74 (66%), Gaps = 1/74 (1%)
Query 218 APASSSQGRGKTSRCHKCNKKVGLLGFVCRCGFAFCGEHRYADAHGCVFDYKAFEREQLR 277
AP +++ +G SRC CNK+VGL GF CRCG FCG HRYAD H C F+Y A +E +
Sbjct 101 APEEAAKPKGP-SRCTTCNKRVGLTGFKCRCGSLFCGTHRYADVHDCSFNYHAAAQEAIA 159
Query 278 KQNNKVVADKLQKL 291
K N V A+KL K+
Sbjct 160 KANPVVKAEKLDKI 173
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 19/41 (46%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 37 PSAPPLCAKNCGFYGSPANRNLCSKCYREFLKAESAAATAA 77
P P LC NCGF+GS A N+CSKC+++ L + A A
Sbjct 12 PEGPKLCTNNCGFFGSAATMNMCSKCHKDMLFQQEQGAKFA 52
> Hs11345484
Length=227
Score = 67.4 bits (163), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 66/243 (27%), Positives = 91/243 (37%), Gaps = 55/243 (22%)
Query 37 PSAPPLCAKNCGFYGSPANRNLCSKCYREFLKAESAAATAAAACRRALPQQQQNSDAVAG 96
PS PP C CGF+GS NLCSKC+ +F K + +A + SD +
Sbjct 12 PSLPPRCP--CGFWGSSKTMNLCSKCFADFQKKQPDDDSAPST-------SNSQSDLFSE 62
Query 97 DTSSAVPAEQSSAARSQLKAVTEPSDAIGPAAVAATEATAAAATVAAESCGEEATAPTQE 156
+T+S + S PS P + T + + E CG
Sbjct 63 ETTS------DNNNTSITTPTLSPSQQPLPTELNVT-------SPSKEECG--------- 100
Query 157 PADDAAAVGLAPAAVPSTASDPQPGVAATPEATAAAAAATSMTSSSPVESTDTAAPPSPS 216
P D A V L TP + + S +SPV+ S
Sbjct 101 PCTDTAHVSLI-----------------TPTKRSCGTDSQSENEASPVKRPRLLENTERS 143
Query 217 SAPASSSQGRGKTSRCHKCNKKVGL----LGFVCRCGFAFCGEHRYADAHGCVFDYKAFE 272
+ S Q + RC +C K+ L LG CRCG+ FC HR + H C FD+
Sbjct 144 EETSRSKQKSRR--RCFQCQTKLELVQQELG-SCRCGYVFCMLHRLPEQHDCTFDHMGRG 200
Query 273 REQ 275
RE+
Sbjct 201 REE 203
> Hs14741660_2
Length=98
Score = 63.2 bits (152), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 34/78 (43%), Positives = 43/78 (55%), Gaps = 3/78 (3%)
Query 215 PSSAPASSSQGRGKTSRCHKCNKKVGLLG-FVCRCGFAFCGEHRYADAHGCVFDYKAFER 273
P AP + + T+ C C KK GL + CRCG FC HRYA+ HGC +DYK+ R
Sbjct 23 PVKAPLQTK--KKTTNHCFLCGKKTGLASSYECRCGNNFCASHRYAETHGCTYDYKSAGR 80
Query 274 EQLRKQNNKVVADKLQKL 291
L + N V A KL K+
Sbjct 81 RYLHEANPVVNAPKLPKI 98
> ECU04g0270
Length=156
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 26/61 (42%), Positives = 36/61 (59%), Gaps = 1/61 (1%)
Query 232 CHKCNKKVGLLG-FVCRCGFAFCGEHRYADAHGCVFDYKAFEREQLRKQNNKVVADKLQK 290
C KC K++ L + CRCG +C HR+ D H C FDYKA +L QN K++ K+ +
Sbjct 96 CGKCGKRLKLTNNYSCRCGNVYCIRHRFYDQHSCTFDYKAIAIPKLSAQNPKIIGKKIGE 155
Query 291 L 291
L
Sbjct 156 L 156
> Hs18589968
Length=167
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 36/64 (56%), Gaps = 0/64 (0%)
Query 228 KTSRCHKCNKKVGLLGFVCRCGFAFCGEHRYADAHGCVFDYKAFEREQLRKQNNKVVADK 287
K +RC C KKVGL G F +D H C +DYKA ++RK+N VVA+K
Sbjct 104 KKNRCFMCRKKVGLTGLTADVEICFVDFTVNSDKHNCPYDYKAEAAAKIRKENPVVVAEK 163
Query 288 LQKL 291
+Q++
Sbjct 164 IQRI 167
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 18/40 (45%), Positives = 25/40 (62%), Gaps = 1/40 (2%)
Query 31 QHENERPSAPPLCAKNCGFYGSPANRNLCSKCYREFLKAE 70
Q N+ P P LC+ CGFYG+P +CS CY+E L+ +
Sbjct 3 QETNQTP-GPMLCSTGCGFYGNPRTNGMCSVCYKEHLQRQ 41
> CE01016
Length=341
Score = 50.1 bits (118), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 32/58 (55%), Gaps = 2/58 (3%)
Query 228 KTSRCHKCNKKVG--LLGFVCRCGFAFCGEHRYADAHGCVFDYKAFEREQLRKQNNKV 283
+ ++C+ C KK+ C+C FC HR+ H CV DYK R +L+K N+KV
Sbjct 272 RKTKCNTCFKKLSAAQQTMHCKCLRIFCDRHRHPKNHTCVIDYKQDGRNKLKKNNSKV 329
> HsM9506853
Length=186
Score = 46.6 bits (109), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 32 HENERPSAPPLCAKNCGFYGSPANRNLCSKCYREFLKAESAA 73
E P LC+ CGFYG+P +CS CY+E L+ ++++
Sbjct 3 QETNHSQVPMLCSTGCGFYGNPRTNGMCSVCYKEHLQRQNSS 44
> 7299162
Length=97
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 29/54 (53%), Gaps = 2/54 (3%)
Query 35 ERPSAP--PLCAKNCGFYGSPANRNLCSKCYREFLKAESAAATAAAACRRALPQ 86
ER S P P+C CGFYG+PA LCS CY++ L+ + ++ PQ
Sbjct 2 ERESNPMQPMCRSGCGFYGNPATDGLCSVCYKDSLRKKQQPPVSSTPVSVPSPQ 55
> Hs14739640
Length=159
Score = 43.1 bits (100), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 33 ENERPSAPPLCAKNCGFYGSPANRNLCSKCYREFLK 68
E P LC+ CGFYG+P CS CY+E L+
Sbjct 4 ETNHSQVPMLCSTGCGFYGNPRTNGTCSVCYKEHLQ 39
Lambda K H
0.310 0.121 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6460838058
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40