bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2327_orf1
Length=118
Score E
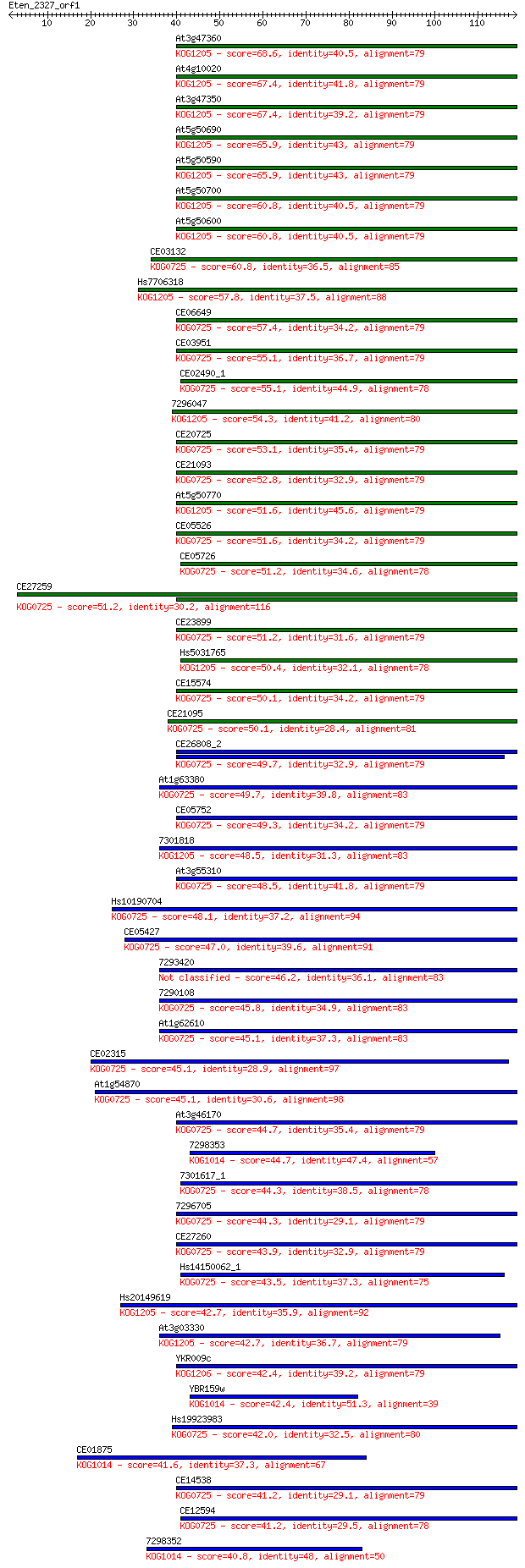
Sequences producing significant alignments: (Bits) Value
At3g47360 68.6 3e-12
At4g10020 67.4 5e-12
At3g47350 67.4 6e-12
At5g50690 65.9 2e-11
At5g50590 65.9 2e-11
At5g50700 60.8 6e-10
At5g50600 60.8 6e-10
CE03132 60.8 6e-10
Hs7706318 57.8 5e-09
CE06649 57.4 7e-09
CE03951 55.1 3e-08
CE02490_1 55.1 3e-08
7296047 54.3 6e-08
CE20725 53.1 1e-07
CE21093 52.8 2e-07
At5g50770 51.6 3e-07
CE05526 51.6 3e-07
CE05726 51.2 4e-07
CE27259 51.2 4e-07
CE23899 51.2 5e-07
Hs5031765 50.4 7e-07
CE15574 50.1 9e-07
CE21095 50.1 1e-06
CE26808_2 49.7 1e-06
At1g63380 49.7 1e-06
CE05752 49.3 2e-06
7301818 48.5 3e-06
At3g55310 48.5 3e-06
Hs10190704 48.1 4e-06
CE05427 47.0 9e-06
7293420 46.2 2e-05
7290108 45.8 2e-05
At1g62610 45.1 3e-05
CE02315 45.1 3e-05
At1g54870 45.1 3e-05
At3g46170 44.7 4e-05
7298353 44.7 4e-05
7301617_1 44.3 5e-05
7296705 44.3 5e-05
CE27260 43.9 6e-05
Hs14150062_1 43.5 1e-04
Hs20149619 42.7 1e-04
At3g03330 42.7 1e-04
YKR009c 42.4 2e-04
YBR159w 42.4 2e-04
Hs19923983 42.0 3e-04
CE01875 41.6 3e-04
CE14538 41.2 4e-04
CE12594 41.2 5e-04
7298352 40.8 6e-04
> At3g47360
Length=309
Score = 68.6 bits (166), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 32/79 (40%), Positives = 44/79 (55%), Gaps = 0/79 (0%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAGGCLAVPTDVTK 99
VVLITGA+ GIG+ +A YA++G L L AR L+ G+G + +P DV+
Sbjct 49 VVLITGASSGIGEHVAYEYAKKGAYLALVARRRDRLEIVAETSRQLGSGNVIIIPGDVSN 108
Query 100 EAACAHLISEAVREFGRLD 118
C I E +R FG+LD
Sbjct 109 VEDCKKFIDETIRHFGKLD 127
> At4g10020
Length=389
Score = 67.4 bits (163), Expect = 5e-12, Method: Composition-based stats.
Identities = 33/79 (41%), Positives = 49/79 (62%), Gaps = 0/79 (0%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAGGCLAVPTDVTK 99
VV+ITGA+ IG+++A YA+RG +LVL AR E+ L+ + GA + + DV K
Sbjct 51 VVVITGASSAIGEQIAYEYAKRGANLVLVARREQRLRVVSNKAKQIGANHVIIIAADVIK 110
Query 100 EAACAHLISEAVREFGRLD 118
E C I++AV +GR+D
Sbjct 111 EDDCRRFITQAVNYYGRVD 129
> At3g47350
Length=308
Score = 67.4 bits (163), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 31/79 (39%), Positives = 44/79 (55%), Gaps = 0/79 (0%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAGGCLAVPTDVTK 99
VVLITGA+ GIG+ +A YA++G L L AR + L+ G+G + +P DV+
Sbjct 48 VVLITGASSGIGEHVAYEYAKKGAKLALVARRKDRLEIVAETSRQLGSGDVIIIPGDVSN 107
Query 100 EAACAHLISEAVREFGRLD 118
C I E + FG+LD
Sbjct 108 VEDCKKFIDETIHHFGKLD 126
> At5g50690
Length=303
Score = 65.9 bits (159), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 34/79 (43%), Positives = 42/79 (53%), Gaps = 0/79 (0%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAGGCLAVPTDVTK 99
VV+ITG++ GIG+ LA YARRG L L AR E LQ C G+ V DV+
Sbjct 49 VVIITGSSSGIGEHLAYEYARRGAYLTLVARREDRLQVVADRCRKLGSPDVAVVRGDVSV 108
Query 100 EAACAHLISEAVREFGRLD 118
C + E + FGRLD
Sbjct 109 IKDCKRFVQETISRFGRLD 127
> At5g50590
Length=299
Score = 65.9 bits (159), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 34/79 (43%), Positives = 42/79 (53%), Gaps = 0/79 (0%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAGGCLAVPTDVTK 99
VV+ITG++ GIG+ LA YARRG L L AR E LQ C G+ V DV+
Sbjct 49 VVIITGSSSGIGEHLAYEYARRGAYLTLVARREDRLQVVADRCRKLGSPDVAVVRGDVSV 108
Query 100 EAACAHLISEAVREFGRLD 118
C + E + FGRLD
Sbjct 109 IKDCKRFVQETISRFGRLD 127
> At5g50700
Length=349
Score = 60.8 bits (146), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 32/79 (40%), Positives = 44/79 (55%), Gaps = 0/79 (0%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAGGCLAVPTDVTK 99
VVLITGA+ GIG++LA YA RG L L AR + L+ G+ + V DV+K
Sbjct 49 VVLITGASSGIGEQLAYEYACRGACLALTARRKNRLEEVAEIARELGSPNVVTVHADVSK 108
Query 100 EAACAHLISEAVREFGRLD 118
C ++ + + FGRLD
Sbjct 109 PDDCRRIVDDTITHFGRLD 127
> At5g50600
Length=349
Score = 60.8 bits (146), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 32/79 (40%), Positives = 44/79 (55%), Gaps = 0/79 (0%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAGGCLAVPTDVTK 99
VVLITGA+ GIG++LA YA RG L L AR + L+ G+ + V DV+K
Sbjct 49 VVLITGASSGIGEQLAYEYACRGACLALTARRKNRLEEVAEIARELGSPNVVTVHADVSK 108
Query 100 EAACAHLISEAVREFGRLD 118
C ++ + + FGRLD
Sbjct 109 PDDCRRIVDDTITHFGRLD 127
> CE03132
Length=278
Score = 60.8 bits (146), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 31/87 (35%), Positives = 48/87 (55%), Gaps = 2/87 (2%)
Query 34 KYIPHSVVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAG--AGGCL 91
K V +ITG++ GIG+E AL +A+ G + + R E+ L+ K L AG L
Sbjct 3 KRFSKKVAIITGSSSGIGRETALLFAKEGAKVTVTGRSEEKLEETKKALLDAGIKESNFL 62
Query 92 AVPTDVTKEAACAHLISEAVREFGRLD 118
VP D+T LIS+ +++FGR++
Sbjct 63 IVPADITFSTGQDELISQTLKKFGRIN 89
> Hs7706318
Length=339
Score = 57.8 bits (138), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 33/91 (36%), Positives = 49/91 (53%), Gaps = 3/91 (3%)
Query 31 RSDKYIPHSVVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAG---A 87
R + + VV +TGA+ GIG+ELA + ++ G SLVL+AR L+ K CL G
Sbjct 43 RPEWELTDMVVWVTGASSGIGEELAYQLSKLGVSLVLSARRVHELERVKRRCLENGNLKE 102
Query 88 GGCLAVPTDVTKEAACAHLISEAVREFGRLD 118
L +P D+T + ++EFGR+D
Sbjct 103 KDILVLPLDLTDTGSHEAATKAVLQEFGRID 133
> CE06649
Length=284
Score = 57.4 bits (137), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 46/81 (56%), Gaps = 2/81 (2%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAGGCLAVPT--DV 97
VVLI+G++KGIG+ A+++A G +VL R ++ + C+ GA +PT D+
Sbjct 10 VVLISGSSKGIGQATAVKFAAEGAKIVLNGRSADDVEKTRKLCMEVGAKPWDLLPTVGDI 69
Query 98 TKEAACAHLISEAVREFGRLD 118
T E +++ + FG+LD
Sbjct 70 TNEDFVKMMVNTVIHNFGKLD 90
> CE03951
Length=277
Score = 55.1 bits (131), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 47/81 (58%), Gaps = 2/81 (2%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGA--GGCLAVPTDV 97
V +ITG++ GIG+ AL +A+ G + + R E+ LQ +K L AG L VP D+
Sbjct 9 VAIITGSSFGIGRATALLFAKEGAKVTVTGRSEERLQGSKQALLDAGISDSNFLIVPADI 68
Query 98 TKEAACAHLISEAVREFGRLD 118
T + LIS+ + +FG+++
Sbjct 69 TTSSGQDALISKTLEKFGQIN 89
> CE02490_1
Length=302
Score = 55.1 bits (131), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 35/85 (41%), Positives = 51/85 (60%), Gaps = 8/85 (9%)
Query 41 VLITGATKGIGKELALRYARRGCSLVLAARGEKA-------LQAAKAECLAAGAGGCLAV 93
VLITGA++GIGKE+AL+ A+ G ++V+AA+ A + +A E AG G L
Sbjct 12 VLITGASRGIGKEIALKLAKDGANIVVAAKTATAHPKLPGTIYSAAEEIEKAG-GKALPC 70
Query 94 PTDVTKEAACAHLISEAVREFGRLD 118
DV EA+ + EAV++FG +D
Sbjct 71 IVDVRDEASVKASVEEAVKKFGGID 95
> 7296047
Length=321
Score = 54.3 bits (129), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 33/85 (38%), Positives = 44/85 (51%), Gaps = 5/85 (5%)
Query 39 SVVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAG-----GCLAV 93
VV ITGA+ GIG+ LAL AR G LVL+AR + L+ + ECLAA G L +
Sbjct 47 QVVWITGASSGIGRALALSLARHGVKLVLSARRLEQLEQVQEECLAAARGLLATKDVLVI 106
Query 94 PTDVTKEAACAHLISEAVREFGRLD 118
D+ ++ + F RLD
Sbjct 107 QMDMLDLDEHKTHLNTVLNHFHRLD 131
> CE20725
Length=279
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 46/81 (56%), Gaps = 2/81 (2%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAG--AGGCLAVPTDV 97
V LITG++ GIG+ A+ +A++G + + R + L+ + E +G A LA+ DV
Sbjct 8 VALITGSSNGIGRAAAILFAQQGAKVTITGRNAERLKETRHEIKKSGIPAENILAIVADV 67
Query 98 TKEAACAHLISEAVREFGRLD 118
+ LI++ VR+FG LD
Sbjct 68 ITDEGQMRLINDTVRKFGHLD 88
> CE21093
Length=281
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 46/81 (56%), Gaps = 2/81 (2%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGA--GGCLAVPTDV 97
V L+TG++ GIG+ A+ +A+ G + + R + L+ + E L +G L+V TD+
Sbjct 9 VALVTGSSNGIGRAAAVLFAKDGAKVTVTGRNAERLEETRQEILKSGVPESHVLSVATDL 68
Query 98 TKEAACAHLISEAVREFGRLD 118
E L++ +++FGRLD
Sbjct 69 AAEKGQDELVNSTIQKFGRLD 89
> At5g50770
Length=342
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 36/81 (44%), Positives = 42/81 (51%), Gaps = 4/81 (4%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLA-ARGEKALQ-AAKAECLAAGAGGCLAVPTDV 97
VV+ITGA GIG+ LA Y +RG L L RGE AA AE G+ L + DV
Sbjct 49 VVVITGAASGIGEALAYEYGKRGAYLALVDIRGEPLFHVAALAELY--GSPEVLPLVADV 106
Query 98 TKEAACAHLISEAVREFGRLD 118
+K C I V FGRLD
Sbjct 107 SKLQDCERFIRATVLHFGRLD 127
> CE05526
Length=278
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 46/81 (56%), Gaps = 2/81 (2%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAG--GCLAVPTDV 97
V +ITG++ GIG+ A+ +AR G + + R + L+ + + LAAG +V DV
Sbjct 8 VAIITGSSNGIGRATAVLFAREGAKVTITGRHAERLEETRQQILAAGVSEQNVNSVVADV 67
Query 98 TKEAACAHLISEAVREFGRLD 118
T +A ++S + +FG+LD
Sbjct 68 TTDAGQDEILSTTLGKFGKLD 88
> CE05726
Length=277
Score = 51.2 bits (121), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 46/80 (57%), Gaps = 2/80 (2%)
Query 41 VLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAG--AGGCLAVPTDVT 98
V+ITG++ GIG+ A+ +A+ G + + R E L+ K + L AG A AV DVT
Sbjct 9 VIITGSSNGIGRSAAVIFAKEGAQVTITGRNEDRLEETKQQILKAGVPAEKINAVVADVT 68
Query 99 KEAACAHLISEAVREFGRLD 118
+ + +I+ + +FG++D
Sbjct 69 EASGQDDIINTTLAKFGKID 88
> CE27259
Length=501
Score = 51.2 bits (121), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 35/119 (29%), Positives = 59/119 (49%), Gaps = 3/119 (2%)
Query 3 VTLQPRSRPRANFAFSSPMGNSPALLALRSDKYIPHSVVLITGATKGIGKELALRYARRG 62
VT R+ + +S+ + S AL +Y V +ITGA+ GIGK AL +A++
Sbjct 186 VTNLHRTSGQDETTYSAFLERSKTTHALGRPEYPKMPVAIITGASSGIGKGTALLFAKKK 245
Query 63 CSLVLAARGEKALQAAKAECLAAG---AGGCLAVPTDVTKEAACAHLISEAVREFGRLD 118
L L R +L+ A C++ G A L +++ + A ++ V++FGR+D
Sbjct 246 YQLSLTGRNTDSLKEVAALCISEGAISADDILITAVELSSDEAPKAIVDATVQKFGRID 304
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 41/81 (50%), Gaps = 2/81 (2%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGA--GGCLAVPTDV 97
V ++TGA+ GIG+ AL AR L L R AL + ++AG+ L ++
Sbjct 3 VAIVTGASSGIGRAAALLLARNNYKLSLTGRNVAALNELAGQIVSAGSDKNDVLVTAVEL 62
Query 98 TKEAACAHLISEAVREFGRLD 118
E A ++ V++FGR+D
Sbjct 63 ASEEAPKTIVDATVQKFGRID 83
> CE23899
Length=279
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 44/81 (54%), Gaps = 2/81 (2%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGA--GGCLAVPTDV 97
V L+TG++ GIG+ A+ A+ G + + R L+ + E L +G L++ TD+
Sbjct 8 VALVTGSSNGIGRATAVLLAQEGAKVTITGRNADRLEETRQEILKSGVPEDHVLSIATDL 67
Query 98 TKEAACAHLISEAVREFGRLD 118
E L++ +++FGRLD
Sbjct 68 ATEKGQDELVNSTIQKFGRLD 88
> Hs5031765
Length=292
Score = 50.4 bits (119), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 25/78 (32%), Positives = 42/78 (53%), Gaps = 0/78 (0%)
Query 41 VLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAGGCLAVPTDVTKE 100
V++TGA+KGIG+E+A A+ G +V+ AR ++ LQ + CL GA + +
Sbjct 37 VIVTGASKGIGREMAYHLAKMGAHVVVTARSKETLQKVVSHCLELGAASAHYIAGTMEDM 96
Query 101 AACAHLISEAVREFGRLD 118
+++A + G LD
Sbjct 97 TFAEQFVAQAGKLMGGLD 114
> CE15574
Length=279
Score = 50.1 bits (118), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 43/81 (53%), Gaps = 2/81 (2%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGA--GGCLAVPTDV 97
V L+TG++ GIG+ A+ AR G + + R + L+ K E L +G L++ D+
Sbjct 8 VALVTGSSNGIGRATAILLAREGAKVTITGRNAQRLEETKQEILRSGVPEDHVLSIIADL 67
Query 98 TKEAACAHLISEAVREFGRLD 118
E+ L++ V FGRLD
Sbjct 68 ATESGQIELMNSTVDIFGRLD 88
> CE21095
Length=280
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 46/83 (55%), Gaps = 2/83 (2%)
Query 38 HSVVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAG--GCLAVPT 95
+ V+ITG++ GIG+ A+ +A+ G ++ + R + L+ + L +G +V
Sbjct 6 NKTVIITGSSNGIGRTTAILFAQEGANVTITGRSSERLEETRQIILKSGVSEKQVNSVVA 65
Query 96 DVTKEAACAHLISEAVREFGRLD 118
DVT E +I+ +++FG++D
Sbjct 66 DVTTEDGQDQIINSTLKQFGKID 88
> CE26808_2
Length=550
Score = 49.7 bits (117), Expect = 1e-06, Method: Composition-based stats.
Identities = 26/81 (32%), Positives = 46/81 (56%), Gaps = 2/81 (2%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAG--AGGCLAVPTDV 97
V L+TG++ GIG+ AL +A++G + + R + L+ + L +G A LA+ D+
Sbjct 223 VALVTGSSNGIGRAAALLFAQQGAKVTITGRNAERLEETRQAILKSGVPAENVLAIAADL 282
Query 98 TKEAACAHLISEAVREFGRLD 118
+ LI+ +++FGRLD
Sbjct 283 ATDQGQTDLINGTLQKFGRLD 303
Score = 42.7 bits (99), Expect = 2e-04, Method: Composition-based stats.
Identities = 23/78 (29%), Positives = 42/78 (53%), Gaps = 2/78 (2%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAG--AGGCLAVPTDV 97
V L+TG++ GIG+ AL +A++G + + R + L+ + L +G A LA+ TD+
Sbjct 8 VALVTGSSNGIGRAAALLFAQQGAKVTITGRNAERLEETRQAILKSGVPAENVLAIATDL 67
Query 98 TKEAACAHLISEAVREFG 115
+ LI+ ++ G
Sbjct 68 ATDQGQTDLINGTLQNIG 85
> At1g63380
Length=287
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/86 (38%), Positives = 47/86 (54%), Gaps = 3/86 (3%)
Query 36 IPHSVVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAGGCLAV-- 93
+ VVL+TGA+ GIG+E+ L + GC +V AAR L + +E + GA G AV
Sbjct 22 LKDKVVLVTGASSGIGREICLDLCKAGCKIVAAARRVDRLNSLCSEINSFGAIGVQAVAL 81
Query 94 PTDVTKEA-ACAHLISEAVREFGRLD 118
DV+ EA + EA FG++D
Sbjct 82 ELDVSSEADTIRKAVKEAWETFGKID 107
> CE05752
Length=284
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 43/81 (53%), Gaps = 2/81 (2%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGA--GGCLAVPTDV 97
V +ITG++ GIG+ A A G + + R + L+ K L AG G L V D+
Sbjct 8 VAIITGSSNGIGQATARLLASEGAKVTVTGRNAERLEETKNILLGAGVPEGNVLVVVGDI 67
Query 98 TKEAACAHLISEAVREFGRLD 118
T+E+ +LI + +FG++D
Sbjct 68 TQESVQENLIKSTLDKFGKID 88
> 7301818
Length=326
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 46/86 (53%), Gaps = 3/86 (3%)
Query 36 IPHSVVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAGGCL---A 92
+P VVLITGA+ G+G+ LA + R GC ++LAAR + L+ K + LA
Sbjct 51 LPGKVVLITGASSGLGESLAHVFYRAGCRVILAARRTQELERVKKDLLALDVDPAYPPTV 110
Query 93 VPTDVTKEAACAHLISEAVREFGRLD 118
+P D+ + + ++ + + ++D
Sbjct 111 LPLDLAELNSIPEFVTRVLAVYNQVD 136
> At3g55310
Length=298
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 33/85 (38%), Positives = 50/85 (58%), Gaps = 9/85 (10%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAGGCL--AVPTDV 97
VVL+TGA+ GIG+E+ L A+ GC ++ AAR L + +E + + G A+ DV
Sbjct 40 VVLVTGASSGIGREICLDLAKAGCQVIAAARRVDRLNSLCSEINSFSSTGIQAAALELDV 99
Query 98 TKEAACAHLISEAVRE----FGRLD 118
+ +AA I +AVRE FG++D
Sbjct 100 SSDAAT---IQKAVREAWDIFGKID 121
> Hs10190704
Length=292
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 35/97 (36%), Positives = 48/97 (49%), Gaps = 6/97 (6%)
Query 25 PALLALRSDKYIPHSVVLITGATKGIGKELALRYARRGCSLVLAARG-EKALQAAKAECL 83
PA L + V ITG GIG +A + R GC V+A+R + L AA+
Sbjct 15 PAYRHLFCPDLLRDKVAFITGGGSGIGFRIAEIFMRHGCHTVIASRSLPRVLTAARK--- 71
Query 84 AAGAGG--CLAVPTDVTKEAACAHLISEAVREFGRLD 118
AGA G CL + DV A + +A++EFGR+D
Sbjct 72 LAGATGRRCLPLSMDVRAPPAVMAAVDQALKEFGRID 108
> CE05427
Length=293
Score = 47.0 bits (110), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 36/98 (36%), Positives = 55/98 (56%), Gaps = 8/98 (8%)
Query 28 LALRSDKYIPHSVVLITGATKGIGKELALRYARRGCSLVLAARGEKA-------LQAAKA 80
+A+++ + V ITGA++GIGKE+AL+ A+ G ++V+AA+ A + A A
Sbjct 3 VAIKNTGFFAGKTVFITGASRGIGKEIALKLAKDGANIVVAAKTATAHPKLPGTIYTAAA 62
Query 81 ECLAAGAGGCLAVPTDVTKEAACAHLISEAVREFGRLD 118
E AG G L DV EAA + AV++FG +D
Sbjct 63 EIEKAG-GHALPCVVDVRDEAAVKAAVDAAVKKFGGID 99
> 7293420
Length=255
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/84 (35%), Positives = 46/84 (54%), Gaps = 6/84 (7%)
Query 36 IPHSVVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAGG-CLAVP 94
I ++V L+TG G+G+ A R A++G S++LA L ++K +A G + VP
Sbjct 2 IKNAVSLVTGGASGLGRATAERLAKQGASVILA-----DLPSSKGNEVAKELGDKVVFVP 56
Query 95 TDVTKEAACAHLISEAVREFGRLD 118
DVT E + + A +FGRLD
Sbjct 57 VDVTSEKDVSAALQTAKDKFGRLD 80
> 7290108
Length=270
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 29/83 (34%), Positives = 45/83 (54%), Gaps = 5/83 (6%)
Query 36 IPHSVVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAGGCLAVPT 95
+ + VV++TGA+ GIG +A AR G +L L R L+A K A +A
Sbjct 3 LSNKVVIVTGASSGIGAAIAQVLAREGATLALVGRNVANLEATKKSLKGTQAEIVVA--- 59
Query 96 DVTKEAACAHLISEAVREFGRLD 118
DVTK+A ++ + + +FGR+D
Sbjct 60 DVTKDADA--IVQQTLAKFGRID 80
> At1g62610
Length=307
Score = 45.1 bits (105), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 31/86 (36%), Positives = 46/86 (53%), Gaps = 3/86 (3%)
Query 36 IPHSVVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAGGCL--AV 93
+ VVL+TGA+ GIG+E+ L + GC +V AAR L + +E + GA G A+
Sbjct 44 LKDKVVLVTGASSGIGREICLDLCKAGCKIVAAARRVDRLNSLCSEINSFGAIGVQAAAL 103
Query 94 PTDVTKEA-ACAHLISEAVREFGRLD 118
DV+ +A + EA FG +D
Sbjct 104 ELDVSSDADTIRKAVKEAWEIFGTID 129
> CE02315
Length=309
Score = 45.1 bits (105), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 44/97 (45%), Gaps = 4/97 (4%)
Query 20 PMGNSPALLALRSDKYIPHSVVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAK 79
P+ NSP L D + V L+TG GIGK +A +A G S+ +AAR + L+
Sbjct 11 PICNSPIL----RDGALKGKVALVTGGGTGIGKAIATTFAHLGASVAIAARRMEKLEQTA 66
Query 80 AECLAAGAGGCLAVPTDVTKEAACAHLISEAVREFGR 116
E + G C D+ + + ++FG+
Sbjct 67 EEIMKTTGGICEPFRMDIKDPGMVSDTFDKIDKKFGK 103
> At1g54870
Length=286
Score = 45.1 bits (105), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 30/107 (28%), Positives = 50/107 (46%), Gaps = 12/107 (11%)
Query 21 MGNSPALLALRSDKYIPHS---VVLITGATKGIGKELALRYARRGCSLVLA-ARGEKALQ 76
M +SP S Y P + V LITG GIG+ + +A G ++ +G++
Sbjct 18 MESSPQF---SSSDYQPSNKLRVALITGGDSGIGRAVGYCFASEGATVAFTYVKGQEEKD 74
Query 77 AAKA-----ECLAAGAGGCLAVPTDVTKEAACAHLISEAVREFGRLD 118
A + E + + +A+PTD+ + C ++ E V FGR+D
Sbjct 75 AQETLQMLKEVKTSDSKEPIAIPTDLGFDENCKRVVDEVVNAFGRID 121
> At3g46170
Length=288
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 28/82 (34%), Positives = 46/82 (56%), Gaps = 3/82 (3%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAGGCL--AVPTDV 97
VVL+TGA+ GIG+E+ L + GC ++ AR L + +E ++ + G A+ DV
Sbjct 30 VVLVTGASSGIGREICLDLGKAGCKIIAVARRVDRLNSLCSEINSSSSTGIQAAALKLDV 89
Query 98 TKEAACAH-LISEAVREFGRLD 118
T +AA ++ A FG++D
Sbjct 90 TSDAATIQKVVQGAWGIFGKID 111
> 7298353
Length=308
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 27/58 (46%), Positives = 33/58 (56%), Gaps = 1/58 (1%)
Query 43 ITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAG-GCLAVPTDVTK 99
+TGA+ GIGKE A AR+ ++VL AR E+ LQA E GAG V D TK
Sbjct 54 VTGASDGIGKEYAKELARQNINVVLIARTEEKLQAVAKEIADCGAGVQTKIVIADFTK 111
> 7301617_1
Length=302
Score = 44.3 bits (103), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 30/85 (35%), Positives = 45/85 (52%), Gaps = 8/85 (9%)
Query 41 VLITGATKGIGKELALRYARRGCSLVLAARGEK-------ALQAAKAECLAAGAGGCLAV 93
+ ITGA++GIGKE+AL+ AR G ++V+AA+ + + +A E AG G
Sbjct 12 LFITGASRGIGKEIALKAARDGANIVVAAKTAEPHPKLPGTIYSAAEEIEKAG-GKAYPC 70
Query 94 PTDVTKEAACAHLISEAVREFGRLD 118
DV E + AV +FG +D
Sbjct 71 VVDVRDEQQVRSAVEAAVAKFGGID 95
> 7296705
Length=257
Score = 44.3 bits (103), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 40/79 (50%), Gaps = 0/79 (0%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAGGCLAVPTDVTK 99
V+++TGA+ GIG ++ A+ G L + R L + +AAG L V D+
Sbjct 8 VIIVTGASSGIGAGTSVLLAKLGGLLTIVGRNLDKLNETAEQIVAAGGAPALQVAADINS 67
Query 100 EAACAHLISEAVREFGRLD 118
E+ ++S + + GR+D
Sbjct 68 ESDVQGIVSATLAKHGRID 86
> CE27260
Length=206
Score = 43.9 bits (102), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 41/81 (50%), Gaps = 2/81 (2%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGA--GGCLAVPTDV 97
V ++TGA+ GIG+ AL AR L L R AL + ++AG+ L ++
Sbjct 3 VAIVTGASSGIGRAAALLLARNNYKLSLTGRNVAALNELAGQIVSAGSDKNDVLVTAVEL 62
Query 98 TKEAACAHLISEAVREFGRLD 118
E A ++ V++FGR+D
Sbjct 63 ASEEAPKTIVDATVQKFGRID 83
> Hs14150062_1
Length=212
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 28/82 (34%), Positives = 45/82 (54%), Gaps = 8/82 (9%)
Query 41 VLITGATKGIGKELALRYARRGCSLVLAARGEK-------ALQAAKAECLAAGAGGCLAV 93
V ITGA++GIGK +AL+ A+ G ++V+AA+ + + A E A G G L
Sbjct 13 VFITGASRGIGKAIALKAAKDGANIVIAAKTAQPHPKLLGTIYTAAEEIEAVG-GKALPC 71
Query 94 PTDVTKEAACAHLISEAVREFG 115
DV E + + +A+++FG
Sbjct 72 IVDVRDEQQISAAVEKAIKKFG 93
> Hs20149619
Length=325
Score = 42.7 bits (99), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/97 (34%), Positives = 47/97 (48%), Gaps = 5/97 (5%)
Query 27 LLALRSDKYIPHSVVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAG 86
L +R Y+ ++VV+ITGAT G+GKE A + G LVL R AL+ E A+
Sbjct 41 LQWVRGKAYLRNAVVVITGATSGLGKECAKVFYAAGAKLVLCGRNGGALEELIRELTASH 100
Query 87 AGGC-----LAVPTDVTKEAACAHLISEAVREFGRLD 118
A V D+T A +E ++ FG +D
Sbjct 101 ATKVQTHKPYLVTFDLTDSGAIVAAAAEILQCFGYVD 137
> At3g03330
Length=345
Score = 42.7 bits (99), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 45/81 (55%), Gaps = 2/81 (2%)
Query 36 IPHSVVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAGGCLAV-P 94
I VV ITGA++GIG+ LA ++A L+L+AR + L K+E A + V P
Sbjct 41 IQGKVVWITGASRGIGEILAKQFASLDAKLILSARNKAELDRVKSELKGKFAPEDVKVLP 100
Query 95 TDV-TKEAACAHLISEAVREF 114
D+ + E + H++ +AV F
Sbjct 101 LDLASGEESLKHVVEQAVSLF 121
> YKR009c
Length=900
Score = 42.4 bits (98), Expect = 2e-04, Method: Composition-based stats.
Identities = 31/86 (36%), Positives = 39/86 (45%), Gaps = 8/86 (9%)
Query 40 VVLITGATKGIGKELALRYARRGCSLV-------LAARGEKALQAAKAECLAAGAGGCLA 92
VV+ITGA G+GK AL YA RG +V L G + A AGG
Sbjct 11 VVVITGAGGGLGKVYALAYASRGAKVVVNDLGGTLGGSGHNSKAADLVVDEIKKAGGIAV 70
Query 93 VPTDVTKEAACAHLISEAVREFGRLD 118
D E +I A++EFGR+D
Sbjct 71 ANYDSVNENG-EKIIETAIKEFGRVD 95
> YBR159w
Length=347
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/39 (51%), Positives = 28/39 (71%), Gaps = 0/39 (0%)
Query 43 ITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAE 81
ITGA+ GIGKE A + A+RG +LVL +R + L+A + E
Sbjct 67 ITGASDGIGKEFARQMAKRGFNLVLISRTQSKLEALQKE 105
> Hs19923983
Length=313
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 44/81 (54%), Gaps = 2/81 (2%)
Query 39 SVVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAGAGGCLAVPTDVT 98
V ++TGA++GIG+ +AL+ + G ++ + R L+ E + G G C+ V D +
Sbjct 8 QVCVVTGASRGIGRGIALQLCKAGATVYITGRHLDTLRVVAQEAQSLG-GQCVPVVCDSS 66
Query 99 KEAACAHLISEAVRE-FGRLD 118
+E+ L + RE GRLD
Sbjct 67 QESEVRSLFEQVDREQQGRLD 87
> CE01875
Length=333
Score = 41.6 bits (96), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 35/67 (52%), Gaps = 0/67 (0%)
Query 17 FSSPMGNSPALLALRSDKYIPHSVVLITGATKGIGKELALRYARRGCSLVLAARGEKALQ 76
FS+ +G L + K S ++TGAT GIGK A ARRG +++L +R + L
Sbjct 43 FSNILGPYVLLSPIDLKKRAGASWAVVTGATDGIGKAYAFELARRGFNVLLVSRTQSKLD 102
Query 77 AAKAECL 83
K E L
Sbjct 103 ETKKEIL 109
> CE14538
Length=280
Score = 41.2 bits (95), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 43/81 (53%), Gaps = 2/81 (2%)
Query 40 VVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAG--AGGCLAVPTDV 97
V ++TG++ GIG+ A+ A G + + R + L+ ++ L G +G +V DV
Sbjct 9 VAIVTGSSNGIGRATAILLASEGAKVTITGRNAERLEESRQALLKVGVPSGHINSVVADV 68
Query 98 TKEAACAHLISEAVREFGRLD 118
T A LI +++FG+++
Sbjct 69 TTGAGQDVLIDSTLKKFGKIN 89
> CE12594
Length=277
Score = 41.2 bits (95), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 43/80 (53%), Gaps = 2/80 (2%)
Query 41 VLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAECLAAG--AGGCLAVPTDVT 98
++ITG++ GIG+ A+ +A+ G + + R L+ + + L AG A AV DV
Sbjct 9 IIITGSSSGIGRSAAVIFAKEGAQVTITGRSADRLEETRLQILKAGVPAEKINAVVADVC 68
Query 99 KEAACAHLISEAVREFGRLD 118
+ + +I + +FG++D
Sbjct 69 EASGQDEVIRTTLAKFGKID 88
> 7298352
Length=339
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 24/50 (48%), Positives = 31/50 (62%), Gaps = 2/50 (4%)
Query 33 DKYIPHSVVLITGATKGIGKELALRYARRGCSLVLAARGEKALQAAKAEC 82
DK+ +VV TGAT GIGKE A AR+G +LVL +R ++ L A E
Sbjct 67 DKFGQWAVV--TGATDGIGKEYARELARQGINLVLISRTKEKLIAVTNEI 114
Lambda K H
0.320 0.133 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167969826
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40