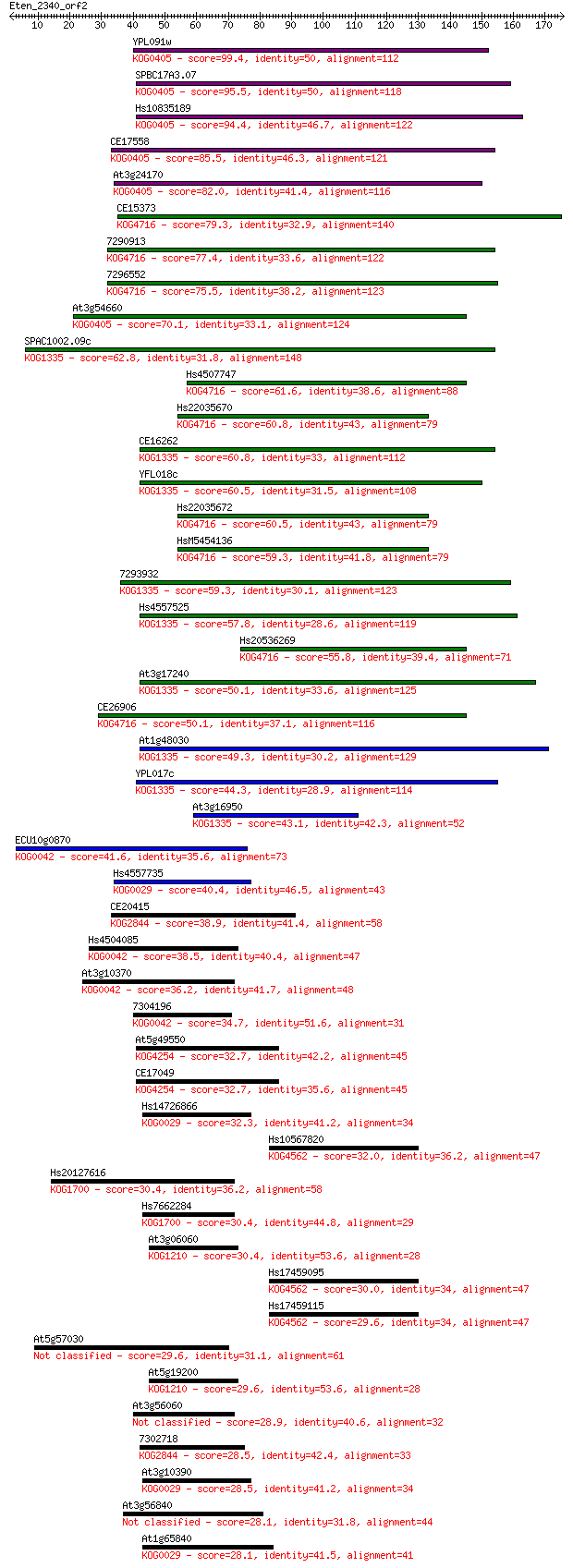
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2340_orf2
Length=175
Score E
Sequences producing significant alignments: (Bits) Value
YPL091w 99.4 3e-21
SPBC17A3.07 95.5 5e-20
Hs10835189 94.4 1e-19
CE17558 85.5 4e-17
At3g24170 82.0 6e-16
CE15373 79.3 4e-15
7290913 77.4 1e-14
7296552 75.5 5e-14
At3g54660 70.1 2e-12
SPAC1002.09c 62.8 3e-10
Hs4507747 61.6 7e-10
Hs22035670 60.8 1e-09
CE16262 60.8 1e-09
YFL018c 60.5 2e-09
Hs22035672 60.5 2e-09
HsM5454136 59.3 4e-09
7293932 59.3 4e-09
Hs4557525 57.8 1e-08
Hs20536269 55.8 4e-08
At3g17240 50.1 2e-06
CE26906 50.1 3e-06
At1g48030 49.3 4e-06
YPL017c 44.3 1e-04
At3g16950 43.1 3e-04
ECU10g0870 41.6 0.001
Hs4557735 40.4 0.002
CE20415 38.9 0.005
Hs4504085 38.5 0.006
At3g10370 36.2 0.031
7304196 34.7 0.088
At5g49550 32.7 0.35
CE17049 32.7 0.42
Hs14726866 32.3 0.51
Hs10567820 32.0 0.70
Hs20127616 30.4 1.9
Hs7662284 30.4 1.9
At3g06060 30.4 2.0
Hs17459095 30.0 2.3
Hs17459115 29.6 3.2
At5g57030 29.6 3.2
At5g19200 29.6 3.3
At3g56060 28.9 5.8
7302718 28.5 7.1
At3g10390 28.5 7.5
At3g56840 28.1 8.2
At1g65840 28.1 8.2
> YPL091w
Length=483
Score = 99.4 bits (246), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 56/118 (47%), Positives = 76/118 (64%), Gaps = 7/118 (5%)
Query 40 HFDCVVLGGGSGGIAFARRAATYGAKVALVERSRMGGTCVNVGCVPKKIMWCAANAFESL 99
H+D +V+GGGSGG+A ARRAA+YGAK LVE +GGTCVNVGCVPKK+MW A++ +
Sbjct 23 HYDYLVIGGGSGGVASARRAASYGAKTLLVEAKALGGTCVNVGCVPKKVMWYASDLATRV 82
Query 100 QGMKHLGVEFSDPP------RLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFA 151
G+ + + P NW Q R+ Y+ RLN IY+ NL+K +V +G+A
Sbjct 83 SHANEYGL-YQNLPLDKEHLTFNWPEFKQKRDAYVHRLNGIYQKNLEKEKVDVVFGWA 139
> SPBC17A3.07
Length=464
Score = 95.5 bits (236), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 59/119 (49%), Positives = 75/119 (63%), Gaps = 1/119 (0%)
Query 41 FDCVVLGGGSGGIAFARRAATYGAKVALVERS-RMGGTCVNVGCVPKKIMWCAANAFESL 99
FD +V+GGGSGG+A ARRAA +GAKVAL+E S R+GGTCVN GCVPKKIMW A+ +
Sbjct 8 FDYLVIGGGSGGLASARRAAKHGAKVALIEASGRLGGTCVNYGCVPKKIMWNIADLVAKM 67
Query 100 QGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATLKPKDE 158
+ K G S +W + + R+ YI RLN IYE N++K V G A+ E
Sbjct 68 KTAKQNGFPNSQLGSFDWGMIKRKRDAYIGRLNGIYERNVNKDGVAYISGHASFVSPTE 126
> Hs10835189
Length=479
Score = 94.4 bits (233), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 57/126 (45%), Positives = 78/126 (61%), Gaps = 5/126 (3%)
Query 41 FDCVVLGGGSGGIAFARRAATYGAKVALVERSRMGGTCVNVGCVPKKIMWCAANAFESLQ 100
+D +V+GGGSGG+A ARRAA GA+ A+VE ++GGTCVNVGCVPKK+MW A E +
Sbjct 22 YDYLVIGGGSGGLASARRAAELGARAAVVESHKLGGTCVNVGCVPKKVMWNTAVHSEFMH 81
Query 101 GMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATL----KPK 156
G S + NW+ + + R+ Y+ RLN IY++NL KS + G A KP
Sbjct 82 DHADYGFP-SCEGKFNWRVIKEKRDAYVSRLNAIYQNNLTKSHIEIIRGHAAFTSDPKPT 140
Query 157 DEANGQ 162
E +G+
Sbjct 141 IEVSGK 146
> CE17558
Length=473
Score = 85.5 bits (210), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 56/122 (45%), Positives = 78/122 (63%), Gaps = 3/122 (2%)
Query 33 SAMAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVERSRMGGTCVNVGCVPKKIMW-C 91
S M+ FD +V+GGGSGGIA ARRA +G V L+E R+GGTCVNVGCVPKK+M+ C
Sbjct 13 SIMSGVKEFDYLVIGGGSGGIASARRAREFGVSVGLIESGRLGGTCVNVGCVPKKVMYNC 72
Query 92 AANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFA 151
+ +A E ++ G + + + +W+ + ++R+ YIKRLN +YE L S V G A
Sbjct 73 SLHA-EFIRDHADYGFDVT-LNKFDWKVIKKSRDEYIKRLNGLYESGLKGSSVEYIRGRA 130
Query 152 TL 153
T
Sbjct 131 TF 132
> At3g24170
Length=499
Score = 82.0 bits (201), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 48/126 (38%), Positives = 67/126 (53%), Gaps = 10/126 (7%)
Query 34 AMAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVER----------SRMGGTCVNVGC 83
A A + FD V+G GSGG+ AR +A +GAKV + E +GGTCV GC
Sbjct 19 ANATHYDFDLFVIGAGSGGVRAARFSANHGAKVGICELPFHPISSEEIGGVGGTCVIRGC 78
Query 84 VPKKIMWCAANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQ 143
VPKKI+ A L+ K+ G E ++ W++L+Q + + I RLN IY+ L +
Sbjct 79 VPKKILVYGATYGGELEDAKNYGWEINEKVDFTWKKLLQKKTDEILRLNNIYKRLLANAA 138
Query 144 VHRYYG 149
V Y G
Sbjct 139 VKLYEG 144
> CE15373
Length=503
Score = 79.3 bits (194), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 46/151 (30%), Positives = 81/151 (53%), Gaps = 16/151 (10%)
Query 35 MAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVERSR---------MGGTCVNVGCVP 85
+ +++ FD +V+G GSGG++ ++RAA GA VAL++ +GGTC NVGC+P
Sbjct 15 LFSSNKFDLIVIGAGSGGLSCSKRAADLGANVALIDAVEPTPHGHSWGIGGTCANVGCIP 74
Query 86 KKIMWCAANAFESLQGMKHLGVEFSDPPRL--NWQRLVQNRENYIKRLNRIYEDNLDKSQ 143
KK+M AA + L+ G D ++ +W L +N + +K N IY L++ +
Sbjct 75 KKLMHQAAIVGKELKHADKYGWNGIDQEKIKHDWNVLSKNVNDRVKANNWIYRVQLNQKK 134
Query 144 VHRYYGFATLKPKDEANGQHVVVVNSSKEEA 174
++ + +A KD+ +V+ + K +
Sbjct 135 INYFNAYAEFVDKDK-----IVITGTDKNKT 160
> 7290913
Length=491
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 41/132 (31%), Positives = 73/132 (55%), Gaps = 10/132 (7%)
Query 32 LSAMAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVERSR----------MGGTCVNV 81
++ + ++ +D +V+GGGS G+A A+ A GA+VA ++ + +GGTCVNV
Sbjct 1 MAPVQGSYDYDLIVIGGGSAGLACAKEAVLNGARVACLDFVKPTPTLGTKWGVGGTCVNV 60
Query 82 GCVPKKIMWCAANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDK 141
GC+PKK+M A+ E++ G + + +W +LVQ+ +N+IK +N + +L
Sbjct 61 GCIPKKLMHQASLLGEAVHEAAAYGWNVDEKIKPDWHKLVQSVQNHIKSVNWVTRVDLRD 120
Query 142 SQVHRYYGFATL 153
+V G +
Sbjct 121 KKVEYINGLGSF 132
> 7296552
Length=516
Score = 75.5 bits (184), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 47/133 (35%), Positives = 73/133 (54%), Gaps = 10/133 (7%)
Query 32 LSAMAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVERSR---------MGGTCVNVG 82
L+A + +D VVLGGGS G+A A+ AA GA+V + + +GGTCVNVG
Sbjct 24 LAASRPRYDYDLVVLGGGSAGLACAKEAAGCGARVLCFDYVKPTPVGTKWGIGGTCVNVG 83
Query 83 CVPKKIMWCAANAFESLQGMKHLGVEFSDP-PRLNWQRLVQNRENYIKRLNRIYEDNLDK 141
C+PKK+M A+ E++ G D R +W++LV++ +N+IK +N + +L
Sbjct 84 CIPKKLMHQASLLGEAVHEAVAYGWNVDDTNIRPDWRKLVRSVQNHIKSVNWVTRVDLRD 143
Query 142 SQVHRYYGFATLK 154
+V AT +
Sbjct 144 KKVEYVNSMATFR 156
> At3g54660
Length=565
Score = 70.1 bits (170), Expect = 2e-12, Method: Composition-based stats.
Identities = 41/134 (30%), Positives = 66/134 (49%), Gaps = 10/134 (7%)
Query 21 QRDSAPASQPHLSAMAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVE---------- 70
+R S AS + + + FD +G GSGG+ +R A ++GA A+ E
Sbjct 68 RRFSVCASTDNGAESDRHYDFDLFTIGAGSGGVRASRFATSFGASAAVCELPFSTISSDT 127
Query 71 RSRMGGTCVNVGCVPKKIMWCAANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIKR 130
+GGTCV GCVPKK++ A+ + G ++ P +W L+ N+ ++R
Sbjct 128 AGGVGGTCVLRGCVPKKLLVYASKYSHEFEDSHGFGWKYETEPSHDWTTLIANKNAELQR 187
Query 131 LNRIYEDNLDKSQV 144
L IY++ L K+ V
Sbjct 188 LTGIYKNILSKANV 201
> SPAC1002.09c
Length=511
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 47/155 (30%), Positives = 77/155 (49%), Gaps = 16/155 (10%)
Query 6 FKFKCCFLSLQQQLQQRDSAPA----SQPHLSAMAAAH-HFDCVVLGGGSGGIAFARRAA 60
FKF C LQ + PA S+ SA A+ + +D V+GGG GG A R A
Sbjct 14 FKFTC--------LQVSECRPAQIEISKRLYSAKASGNGEYDLCVIGGGPGGYVAAIRGA 65
Query 61 TYGAKVALVE-RSRMGGTCVNVGCVPKKIMWCAANAFESLQ-GMKHLGVEFSDPPRLNWQ 118
G K VE R +GGTC+NVGC+P K + ++ + +++ K G++ S +N
Sbjct 66 QLGLKTICVEKRGTLGGTCLNVGCIPSKALLNNSHIYHTVKHDTKRRGIDVSG-VSVNLS 124
Query 119 RLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATL 153
++++ +++ +K L E K++V G +
Sbjct 125 QMMKAKDDSVKSLTSGIEYLFKKNKVEYAKGTGSF 159
> Hs4507747
Length=497
Score = 61.6 bits (148), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 34/97 (35%), Positives = 56/97 (57%), Gaps = 9/97 (9%)
Query 57 RRAATYGAKVALVE---------RSRMGGTCVNVGCVPKKIMWCAANAFESLQGMKHLGV 107
+ AA YG KV +++ R +GGTCVNVGC+PKK+M AA ++LQ ++ G
Sbjct 29 KEAAQYGKKVMVLDFVTPTPLGTRWGLGGTCVNVGCIPKKLMHQAALLGQALQDSRNYGW 88
Query 108 EFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQV 144
+ + + +W R+++ +N+I LN Y L + +V
Sbjct 89 KVEETVKHDWDRMIEAVQNHIGSLNWGYRVALREKKV 125
> Hs22035670
Length=494
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 34/88 (38%), Positives = 51/88 (57%), Gaps = 9/88 (10%)
Query 54 AFARRAATYGAKVALVE---------RSRMGGTCVNVGCVPKKIMWCAANAFESLQGMKH 104
A A+ AA G KVA+V+ R +GGTCVNVGC+PKK+M AA +Q +
Sbjct 23 ACAKEAAQLGRKVAVVDYVEPSPQGTRWGLGGTCVNVGCIPKKLMHQAALLGGLIQDAPN 82
Query 105 LGVEFSDPPRLNWQRLVQNRENYIKRLN 132
G E + P +W+++ + +N++K LN
Sbjct 83 YGWEVAQPVPHDWRKMAEAVQNHVKSLN 110
> CE16262
Length=464
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 37/114 (32%), Positives = 61/114 (53%), Gaps = 4/114 (3%)
Query 42 DCVVLGGGSGGIAFARRAATYGAKVALVER-SRMGGTCVNVGCVPKKIMWCAANAFESLQ 100
D VV+GGG GG A +AA G K VE+ + +GGTC+NVGC+P K + ++ Q
Sbjct 31 DLVVIGGGPGGYVAAIKAAQLGMKTVCVEKNATLGGTCLNVGCIPSKALLNNSHYLHMAQ 90
Query 101 -GMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATL 153
G++ + LN ++++ + N +K+L + ++V GFAT+
Sbjct 91 HDFAARGIDCT--ASLNLPKMMEAKSNSVKQLTGGIKQLFKANKVGHVEGFATI 142
> YFL018c
Length=499
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 34/110 (30%), Positives = 59/110 (53%), Gaps = 2/110 (1%)
Query 42 DCVVLGGGSGGIAFARRAATYGAKVALVE-RSRMGGTCVNVGCVPKKIMWCAANAFESLQ 100
D V++GGG G A +AA G A VE R ++GGTC+NVGC+P K + ++ F +
Sbjct 28 DVVIIGGGPAGYVAAIKAAQLGFNTACVEKRGKLGGTCLNVGCIPSKALLNNSHLFHQMH 87
Query 101 G-MKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYG 149
+ G++ + ++N + +++ +K+L E K++V Y G
Sbjct 88 TEAQKRGIDVNGDIKINVANFQKAKDDAVKQLTGGIELLFKKNKVTYYKG 137
> Hs22035672
Length=524
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 34/88 (38%), Positives = 51/88 (57%), Gaps = 9/88 (10%)
Query 54 AFARRAATYGAKVALVE---------RSRMGGTCVNVGCVPKKIMWCAANAFESLQGMKH 104
A A+ AA G KVA+V+ R +GGTCVNVGC+PKK+M AA +Q +
Sbjct 53 ACAKEAAQLGRKVAVVDYVEPSPQGTRWGLGGTCVNVGCIPKKLMHQAALLGGLIQDAPN 112
Query 105 LGVEFSDPPRLNWQRLVQNRENYIKRLN 132
G E + P +W+++ + +N++K LN
Sbjct 113 YGWEVAQPVPHDWRKMAEAVQNHVKSLN 140
> HsM5454136
Length=524
Score = 59.3 bits (142), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 33/88 (37%), Positives = 51/88 (57%), Gaps = 9/88 (10%)
Query 54 AFARRAATYGAKVALVE---------RSRMGGTCVNVGCVPKKIMWCAANAFESLQGMKH 104
A A+ AA G KV++V+ R +GGTCVNVGC+PKK+M AA +Q +
Sbjct 53 ACAKEAAQLGRKVSVVDYVEPSPQGTRWGLGGTCVNVGCIPKKLMHQAALLGGLIQDAPN 112
Query 105 LGVEFSDPPRLNWQRLVQNRENYIKRLN 132
G E + P +W+++ + +N++K LN
Sbjct 113 YGWEVAQPVPHDWRKMAEAVQNHVKSLN 140
> 7293932
Length=504
Score = 59.3 bits (142), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 64/126 (50%), Gaps = 4/126 (3%)
Query 36 AAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVER-SRMGGTCVNVGCVPKKIMWCAAN 94
++ H D VV+G G GG A +AA G K VE+ + +GGTC+NVGC+P K + ++
Sbjct 33 SSTHEADIVVIGSGPGGYVAAIKAAQMGMKTVSVEKEATLGGTCLNVGCIPSKALLNNSH 92
Query 95 AFESLQG--MKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFAT 152
+ ++ G+ L+ ++L+ + N +K L K++V + GF T
Sbjct 93 YYHMAHSGDLEKRGISCGS-VSLDLEKLMGQKSNAVKALTGGIAMLFKKNKVTQLTGFGT 151
Query 153 LKPKDE 158
+ +E
Sbjct 152 IVNPNE 157
> Hs4557525
Length=509
Score = 57.8 bits (138), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 61/122 (50%), Gaps = 4/122 (3%)
Query 42 DCVVLGGGSGGIAFARRAATYGAKVALVERSR-MGGTCVNVGCVPKKIMWCAANAFESLQ 100
D V+G G GG A +AA G K +E++ +GGTC+NVGC+P K + ++ +
Sbjct 43 DVTVIGSGPGGYVAAIKAAQLGFKTVCIEKNETLGGTCLNVGCIPSKALLNNSHYYHMAH 102
Query 101 GMKHL--GVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATLKPKDE 158
G G+E S+ RLN ++++ + +K L +++V G+ + K++
Sbjct 103 GTDFASRGIEMSE-VRLNLDKMMEQKSTAVKALTGGIAHLFKQNKVVHVNGYGKITGKNQ 161
Query 159 AN 160
Sbjct 162 VT 163
> Hs20536269
Length=459
Score = 55.8 bits (133), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 28/71 (39%), Positives = 43/71 (60%), Gaps = 0/71 (0%)
Query 74 MGGTCVNVGCVPKKIMWCAANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNR 133
+GGTCVNVGC+PKK+M AA ++L + G E++ R NW+ + + +N+I LN
Sbjct 17 LGGTCVNVGCIPKKLMHQAALLGQALCDSRKFGWEYNQQVRHNWETMTKAIQNHISSLNW 76
Query 134 IYEDNLDKSQV 144
Y +L + V
Sbjct 77 GYRLSLREKAV 87
> At3g17240
Length=507
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 42/132 (31%), Positives = 71/132 (53%), Gaps = 8/132 (6%)
Query 42 DCVVLGGGSGGIAFARRAATYGAKVALVE-RSRMGGTCVNVGCVPKKIMWCAANAF-ESL 99
D V++GGG GG A +AA G K +E R +GGTC+NVGC+P K + +++ + E+
Sbjct 45 DVVIIGGGPGGYVAAIKAAQLGLKTTCIEKRGALGGTCLNVGCIPSKALLHSSHMYHEAK 104
Query 100 QGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFAT-LKPK-- 156
+ GV+ S ++ ++ ++ +K L R E K++V+ G+ L P
Sbjct 105 HVFANHGVKVSS-VEVDLPAMLAQKDTAVKNLTRGVEGLFKKNKVNYVKGYGKFLSPSEV 163
Query 157 --DEANGQHVVV 166
D +G++VVV
Sbjct 164 SVDTIDGENVVV 175
> CE26906
Length=665
Score = 50.1 bits (118), Expect = 3e-06, Method: Composition-based stats.
Identities = 43/126 (34%), Positives = 65/126 (51%), Gaps = 10/126 (7%)
Query 29 QPHLSAMAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVERSR---------MGGTCV 79
Q +L H +D +V+GGGSGG+A A+ A+ G KVA ++ + +GGTCV
Sbjct 161 QDYLKEWLRDHTYDLIVIGGGSGGLAAAKEASRLGKKVACLDFVKPSPQGTSWGLGGTCV 220
Query 80 NVGCVPKKIMWCAANAFESLQGMKHLGVEFSDPP-RLNWQRLVQNRENYIKRLNRIYEDN 138
NVGC+PKK+M A+ S+ K G + + W L + +++I LN Y
Sbjct 221 NVGCIPKKLMHQASLLGHSIHDAKKYGWKLPEGKVEHQWNHLRDSVQDHIASLNWGYRVQ 280
Query 139 LDKSQV 144
L + V
Sbjct 281 LREKTV 286
> At1g48030
Length=507
Score = 49.3 bits (116), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 39/143 (27%), Positives = 71/143 (49%), Gaps = 15/143 (10%)
Query 42 DCVVLGGGSGGIAFARRAATYGAKVALVE-RSRMGGTCVNVGCVPKKIMWCAANAF-ESL 99
D V++GGG GG A +A+ G K +E R +GGTC+NVGC+P K + +++ + E+
Sbjct 45 DVVIIGGGPGGYVAAIKASQLGLKTTCIEKRGALGGTCLNVGCIPSKALLHSSHMYHEAK 104
Query 100 QGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATLKPKDEA 159
+ G++ S ++ ++ ++N +K L R E K++V G+ +E
Sbjct 105 HSFANHGIKVSS-VEVDLPAMLAQKDNAVKNLTRGIEGLFKKNKVTYVKGYGKFISPNEV 163
Query 160 N------------GQHVVVVNSS 170
+ G+H++V S
Sbjct 164 SVETIDGGNTIVKGKHIIVATGS 186
> YPL017c
Length=499
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/118 (27%), Positives = 55/118 (46%), Gaps = 5/118 (4%)
Query 41 FDCVVLGGGSGGIAFARRAATYGAKVALVE-RSRMGGTCVNVGCVPKKIMWCAANAFESL 99
+D +V+G G GG A +A+ G A V+ R+ +GG + G VP K + + + L
Sbjct 18 YDVLVIGCGPGGFTAAMQASQAGLLTACVDQRASLGGAYLVDGAVPSKTLLYESYLYRLL 77
Query 100 QG---MKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATLK 154
Q ++ G P + + Q ++ I+ L +Y+ L K+ V Y G A K
Sbjct 78 QQQELIEQRGTRLF-PAKFDMQAAQSALKHNIEELGNVYKRELSKNNVTVYKGTAAFK 134
> At3g16950
Length=564
Score = 43.1 bits (100), Expect = 3e-04, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 29/55 (52%), Gaps = 3/55 (5%)
Query 59 AATYGAKVALVERSRMGGTCVNVGCVPKKIMWCAANAFESLQG---MKHLGVEFS 110
A G K A++E +GGTCVN GCVP K + + LQ MK G++ S
Sbjct 105 AVEKGLKTAIIEGDVVGGTCVNRGCVPSKALLAVSGRMRELQNEHHMKSFGLQVS 159
> ECU10g0870
Length=614
Score = 41.6 bits (96), Expect = 0.001, Method: Composition-based stats.
Identities = 26/76 (34%), Positives = 41/76 (53%), Gaps = 3/76 (3%)
Query 3 SLHFKFKCCFLSLQQQLQQR---DSAPASQPHLSAMAAAHHFDCVVLGGGSGGIAFARRA 59
++ F +K F++ + ++ ++ D PAS+ + + FD VV+GGGS G A
Sbjct 15 AMKFLYKRIFVASRLKMIEKPSEDWEPASREAMIERLRSEVFDLVVVGGGSTGAGCALDG 74
Query 60 ATYGAKVALVERSRMG 75
AT G KVALV+ G
Sbjct 75 ATRGLKVALVDAGDFG 90
> Hs4557735
Length=527
Score = 40.4 bits (93), Expect = 0.002, Method: Composition-based stats.
Identities = 20/44 (45%), Positives = 27/44 (61%), Gaps = 1/44 (2%)
Query 34 AMAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVE-RSRMGG 76
A A H FD VV+GGG G++ A+ YG V ++E R R+GG
Sbjct 7 ASIAGHMFDVVVIGGGISGLSAAKLLTEYGVSVLVLEARDRVGG 50
> CE20415
Length=872
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 24/61 (39%), Positives = 30/61 (49%), Gaps = 3/61 (4%)
Query 33 SAMAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVERSRM---GGTCVNVGCVPKKIM 89
S A + D VV GGG G + A A G KVALVE+ + G T ++ G V I
Sbjct 14 SYTTAVANADVVVCGGGISGTSIAYHLAKRGKKVALVEKDSIGCSGATGLSAGLVSSPIF 73
Query 90 W 90
W
Sbjct 74 W 74
> Hs4504085
Length=727
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 26 PASQPHLSAMAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVERS 72
P+ + L + FD +V+GGG+ G A A T G K ALVER
Sbjct 55 PSREAQLLTLQNTSEFDILVIGGGATGSGCALDAVTRGLKTALVERD 101
> At3g10370
Length=629
Score = 36.2 bits (82), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 26/48 (54%), Gaps = 0/48 (0%)
Query 24 SAPASQPHLSAMAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVER 71
S A + L A A+ D +V+GGG+ G A A T G +V LVER
Sbjct 57 SRSAQESALIAATASDPLDVLVIGGGATGSGVALDAVTRGLRVGLVER 104
> 7304196
Length=713
Score = 34.7 bits (78), Expect = 0.088, Method: Compositional matrix adjust.
Identities = 16/31 (51%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 40 HFDCVVLGGGSGGIAFARRAATYGAKVALVE 70
H+D +++GGG+ G A AAT G K ALVE
Sbjct 54 HYDILIIGGGAVGSGCALDAATRGLKTALVE 84
> At5g49550
Length=647
Score = 32.7 bits (73), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 25/46 (54%), Gaps = 1/46 (2%)
Query 41 FDCVVLGGGSGGIAFARRAATYGAKVALVERSR-MGGTCVNVGCVP 85
+D VV+GGG G+ A A G VA++ER +GG V VP
Sbjct 14 WDAVVIGGGHNGLTAAAYLARGGLSVAVLERRHVIGGAAVTEEIVP 59
> CE17049
Length=544
Score = 32.7 bits (73), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 24/46 (52%), Gaps = 1/46 (2%)
Query 41 FDCVVLGGGSGGIAFARRAATYGAKVALVERSR-MGGTCVNVGCVP 85
+D +++GGG G+ A G KV ++ER +GG V VP
Sbjct 15 YDAIIIGGGHNGLTAAAYLTKAGKKVCVLERRHVVGGAAVTEEIVP 60
> Hs14726866
Length=600
Score = 32.3 bits (72), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 23/35 (65%), Gaps = 1/35 (2%)
Query 43 CVVLGGGSGGIAFARRAATYGAKVALVE-RSRMGG 76
+++G G G+A AR+ ++G V L+E R R+GG
Sbjct 281 VIIIGSGVSGLAAARQLQSFGMDVTLLEARDRVGG 315
> Hs10567820
Length=369
Score = 32.0 bits (71), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 27/47 (57%), Gaps = 4/47 (8%)
Query 83 CVPKKIMWCAANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIK 129
C P++++W A N GM+HL + +P +L Q VQ ENY++
Sbjct 240 CTPEEVIWEALNMMGLYDGMEHLI--YGEPRKLLTQDWVQ--ENYLE 282
> Hs20127616
Length=1067
Score = 30.4 bits (67), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 28/58 (48%), Gaps = 4/58 (6%)
Query 14 SLQQQLQQRDSAPASQPHLSAMAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVER 71
SL +L +R A QP A C+V+G G G+ A A GA+V LVE+
Sbjct 62 SLWTKLDKR----AGQPVYQQGRACTSTKCLVVGAGPCGLRVAVELALLGARVVLVEK 115
> Hs7662284
Length=1124
Score = 30.4 bits (67), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 43 CVVLGGGSGGIAFARRAATYGAKVALVER 71
C+++GGG G+ A A GAKV +VE+
Sbjct 89 CLIVGGGPCGLRTAIELAYLGAKVVVVEK 117
> At3g06060
Length=327
Score = 30.4 bits (67), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 15/29 (51%), Positives = 22/29 (75%), Gaps = 1/29 (3%)
Query 45 VLGGGSG-GIAFARRAATYGAKVALVERS 72
+ GG SG G+A A RAA+ GA+V+++ RS
Sbjct 44 ITGGSSGIGLALAHRAASEGARVSILARS 72
> Hs17459095
Length=307
Score = 30.0 bits (66), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 26/47 (55%), Gaps = 4/47 (8%)
Query 83 CVPKKIMWCAANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIK 129
C P+++MW A N E G +H + +P +L + VQ ENY++
Sbjct 217 CAPEEVMWEALNVIEVYAGREHF--IYGEPRKLLTRDWVQ--ENYLE 259
> Hs17459115
Length=401
Score = 29.6 bits (65), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 26/47 (55%), Gaps = 4/47 (8%)
Query 83 CVPKKIMWCAANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIK 129
C P+++MW A N E G +H + +P +L + VQ ENY++
Sbjct 311 CAPEEVMWEALNVIEVYAGREHF--IYGEPRKLLTRDWVQ--ENYLE 353
> At5g57030
Length=524
Score = 29.6 bits (65), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 30/63 (47%), Gaps = 4/63 (6%)
Query 9 KCCFLSLQQ--QLQQRDSAPASQPHLSAMAAAHHFDCVVLGGGSGGIAFARRAATYGAKV 66
+ F+ +QQ + ++ P +S A D VV+G G G+A A +A G KV
Sbjct 77 EILFVQMQQNKDMDEQSKLVDKLPPISIGDGA--LDLVVIGCGPAGLALAAESAKLGLKV 134
Query 67 ALV 69
L+
Sbjct 135 GLI 137
> At5g19200
Length=331
Score = 29.6 bits (65), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 15/29 (51%), Positives = 21/29 (72%), Gaps = 1/29 (3%)
Query 45 VLGGGSG-GIAFARRAATYGAKVALVERS 72
+ GG SG G+A A RA + GAKV+++ RS
Sbjct 42 ITGGSSGIGLALAHRAVSEGAKVSILARS 70
> At3g56060
Length=557
Score = 28.9 bits (63), Expect = 5.8, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 20/32 (62%), Gaps = 1/32 (3%)
Query 40 HFDCVVLGGGSGGIAFARRAATYGAKVALVER 71
HFD +++GGG+ G A A + A V ++ER
Sbjct 45 HFDYIIIGGGTAGCALA-ATLSQNATVLVLER 75
> 7302718
Length=894
Score = 28.5 bits (62), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 42 DCVVLGGGSGGIAFARRAATYGAKVALVERSRM 74
D VV+GGGS G A G K L+ER+++
Sbjct 38 DVVVIGGGSAGCHTLYHLARRGVKAVLLERAQL 70
> At3g10390
Length=789
Score = 28.5 bits (62), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 22/35 (62%), Gaps = 1/35 (2%)
Query 43 CVVLGGGSGGIAFARRAATYGAKVALVE-RSRMGG 76
+++G G G+A AR+ +G KV ++E R R GG
Sbjct 187 VIIVGAGLSGLAAARQLMRFGFKVTVLEGRKRPGG 221
> At3g56840
Length=483
Score = 28.1 bits (61), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 14/44 (31%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 37 AAHHFDCVVLGGGSGGIAFARRAATYGAKVALVERSRMGGTCVN 80
A D VV+G G G+A AR + G +V +++ + GT +
Sbjct 76 AKERVDTVVIGAGVVGLAVARELSLRGREVLILDAASSFGTVTS 119
> At1g65840
Length=497
Score = 28.1 bits (61), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 25/45 (55%), Gaps = 4/45 (8%)
Query 43 CVVLGGGSGGIAFARRAATYGAKVALVE-RSRMGG---TCVNVGC 83
+V+G G G+A AR + KV ++E R R+GG T + GC
Sbjct 31 VIVIGSGISGLAAARNLSEASFKVTVLESRDRIGGRIHTDYSFGC 75
Lambda K H
0.321 0.133 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2671071884
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40