bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2376_orf1
Length=87
Score E
Sequences producing significant alignments: (Bits) Value
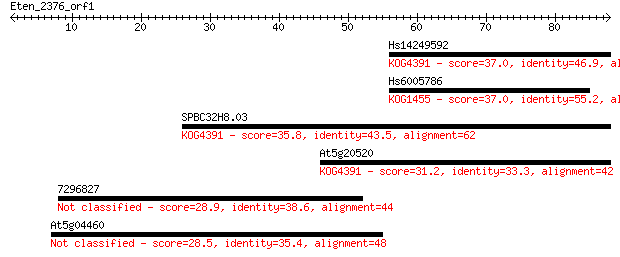
Hs14249592 37.0 0.010
Hs6005786 37.0 0.010
SPBC32H8.03 35.8 0.021
At5g20520 31.2 0.53
7296827 28.9 2.7
At5g04460 28.5 3.2
> Hs14249592
Length=201
Score = 37.0 bits (84), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 56 VFVMGHSMGGAVAIDLAKRRGNELAGLVVENT 87
+F+ G S+GGAVAI LA + ++ ++VENT
Sbjct 51 IFLFGRSLGGAVAIHLASENSHRISAIMVENT 82
> Hs6005786
Length=313
Score = 37.0 bits (84), Expect = 0.010, Method: Composition-based stats.
Identities = 16/29 (55%), Positives = 21/29 (72%), Gaps = 0/29 (0%)
Query 56 VFVMGHSMGGAVAIDLAKRRGNELAGLVV 84
VF++GHSMGGA+AI A R AG+V+
Sbjct 126 VFLLGHSMGGAIAILTAAERPGHFAGMVL 154
> SPBC32H8.03
Length=299
Score = 35.8 bits (81), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 38/80 (47%), Gaps = 19/80 (23%)
Query 26 ANNNNANAISKQQKGKSSSSSSSS------------------SSKSNFVFVMGHSMGGAV 67
A N N IS + GKS+ S S + SK+ V V G S+GGAV
Sbjct 115 ALNMNVFIISYRGYGKSTGSPSEAGLKIDSQTALEYLMEHPICSKTKIV-VYGQSIGGAV 173
Query 68 AIDLAKRRGNELAGLVVENT 87
AI L + + ++ L++ENT
Sbjct 174 AIALTAKNQDRISALILENT 193
> At5g20520
Length=340
Score = 31.2 bits (69), Expect = 0.53, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 46 SSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENT 87
S + ++ + V G S+GGAV L K ++++ L++ENT
Sbjct 170 SGRTDIDTSRIVVFGRSLGGAVGAVLTKNNPDKVSALILENT 211
> 7296827
Length=271
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 17/45 (37%), Positives = 27/45 (60%), Gaps = 1/45 (2%)
Query 8 APQVLHERS-SSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSS 51
A ++ H RS S SNSN NN + N S + K SS++++S++
Sbjct 92 ATRISHRRSIPSQQTSNSNENNQSTNQTSNSRNQKESSTAATSAA 136
> At5g04460
Length=831
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 28/49 (57%), Gaps = 1/49 (2%)
Query 7 GAPQVLHERSSSSSNSNSNANNNN-ANAISKQQKGKSSSSSSSSSSKSN 54
G ++HE +S++ N NSN + N A AI+ + + SS+SS + N
Sbjct 442 GRENIVHENTSNTDNDNSNTSTNALAIAITAGNSQRVTDESSTSSRQGN 490
Lambda K H
0.300 0.115 0.294
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187582654
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40