bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2389_orf1
Length=192
Score E
Sequences producing significant alignments: (Bits) Value
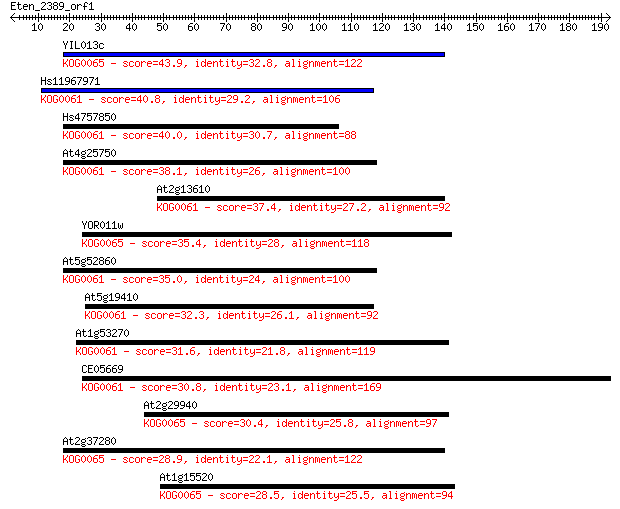
YIL013c 43.9 2e-04
Hs11967971 40.8 0.002
Hs4757850 40.0 0.003
At4g25750 38.1 0.011
At2g13610 37.4 0.018
YOR011w 35.4 0.062
At5g52860 35.0 0.095
At5g19410 32.3 0.64
At1g53270 31.6 1.0
CE05669 30.8 1.6
At2g29940 30.4 2.0
At2g37280 28.9 7.1
At1g15520 28.5 7.6
> YIL013c
Length=1411
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 40/129 (31%), Positives = 58/129 (44%), Gaps = 17/129 (13%)
Query 18 QFKEVGRRAVRLWLRDRSTLMAIMGDSVVQGLVIGAVFF-------GVRSRASLYYQLSA 70
Q K RA + DR+ L++ VVQ LVIG++F+ G SR SL + S
Sbjct 366 QLKTCTVRAFERIIGDRNYLISQFVSVVVQSLVIGSLFYNIPLTTIGSFSRGSLTF-FSI 424
Query 71 LFLMILSLLASSLWTVPLYVQQKAQYRVEVEDGYYSPVPYMFATSLVANFFVLVGDAVLV 130
LF LSL +P Q++ R V+ +Y Y + +L NFF +LV
Sbjct 425 LFFTFLSLA-----DMPASFQRQPVVRKHVQLHFY----YNWVETLATNFFDCCSKFILV 475
Query 131 TIMWLLFGF 139
I ++ F
Sbjct 476 VIFTIILYF 484
> Hs11967971
Length=673
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 31/108 (28%), Positives = 52/108 (48%), Gaps = 2/108 (1%)
Query 11 ELAGPYVQFKEVGRRAVRLWLRDRSTLMAIMGDSVVQGLVIGAVFFGVRSRASLYYQLSA 70
++ G QF + RR + RD TL+ ++ + + IG ++FG S + +A
Sbjct 391 KMPGAVQQFTTLIRRQISNDFRDLPTLLIHGAEACLMSMTIGFLYFGHGSIQLSFMDTAA 450
Query 71 LFLMILSLLASS--LWTVPLYVQQKAQYRVEVEDGYYSPVPYMFATSL 116
L MI +L+ + L + ++A E+EDG Y+ PY FA L
Sbjct 451 LLFMIGALIPFNVILDVISKCYSERAMLYYELEDGLYTTGPYFFAKIL 498
> Hs4757850
Length=655
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 27/89 (30%), Positives = 50/89 (56%), Gaps = 1/89 (1%)
Query 18 QFKEVGRRAVRLWLRDRSTLMAIMGDSVVQGLVIGAVFFGVRSRAS-LYYQLSALFLMIL 76
Q + V +R+ + L + +A + +VV GLVIGA++FG+++ ++ + + LF +
Sbjct 376 QLRWVSKRSFKNLLGNPQASIAQIIVTVVLGLVIGAIYFGLKNDSTGIQNRAGVLFFLTT 435
Query 77 SLLASSLWTVPLYVQQKAQYRVEVEDGYY 105
+ SS+ V L+V +K + E GYY
Sbjct 436 NQCFSSVSAVELFVVEKKLFIHEYISGYY 464
> At4g25750
Length=577
Score = 38.1 bits (87), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 26/104 (25%), Positives = 51/104 (49%), Gaps = 4/104 (3%)
Query 18 QFKEVGRRAVRLW---LRDRSTLMAIMGDSVVQGLVIGAVFFGVRSRASLYYQLSALFLM 74
+ E+ + R W R R L+ + +S+V GLV+G ++ + + + LF
Sbjct 297 RITEISLLSSRFWKIIYRTRQLLLTNILESLVVGLVLGTIYLNIGTGKEGIRKRFGLFAF 356
Query 75 ILS-LLASSLWTVPLYVQQKAQYRVEVEDGYYSPVPYMFATSLV 117
L+ LL+S+ T+P+++ ++ E G Y ++ A +LV
Sbjct 357 TLTFLLSSTTQTLPIFIDERPILLRETSSGLYRLSSHILANTLV 400
> At2g13610
Length=649
Score = 37.4 bits (85), Expect = 0.018, Method: Composition-based stats.
Identities = 25/93 (26%), Positives = 45/93 (48%), Gaps = 1/93 (1%)
Query 48 GLVIGAVFFGVRSRASLYYQLSALFLMILS-LLASSLWTVPLYVQQKAQYRVEVEDGYYS 106
G+V+G +F ++ + LF IL+ LL S++ +P+++Q++ E G Y
Sbjct 402 GIVLGLIFHNLKDDLKGARERVGLFAFILTFLLTSTIEALPIFLQEREILMKETSSGSYR 461
Query 107 PVPYMFATSLVANFFVLVGDAVLVTIMWLLFGF 139
Y A LV F+L+ + T ++ L G
Sbjct 462 VSSYAVANGLVYLPFLLILAILFSTPVYWLVGL 494
> YOR011w
Length=1394
Score = 35.4 bits (80), Expect = 0.062, Method: Composition-based stats.
Identities = 33/125 (26%), Positives = 58/125 (46%), Gaps = 13/125 (10%)
Query 24 RRAVRLWLRDRSTLMAIMGDSVVQGLVIGAVFF-------GVRSRASLYYQLSALFLMIL 76
+RA + L D++ + A V+Q LVIG++F+ G SR SL + S LF L
Sbjct 376 KRAFQRSLGDKAYMTAQFISVVIQSLVIGSLFYEIPLTTIGSYSRGSLTF-FSILFFTFL 434
Query 77 SLLASSLWTVPLYVQQKAQYRVEVEDGYYSPVPYMFATSLVANFFVLVGDAVLVTIMWLL 136
SL +P+ Q++ + + + +Y+ +T++ F L V I++ L
Sbjct 435 SLA-----DMPIAFQRQPVVKKQSQLHFYTNWVETLSTTVFDYCFKLCLVIVFSIILYFL 489
Query 137 FGFQF 141
Q+
Sbjct 490 AHLQY 494
> At5g52860
Length=589
Score = 35.0 bits (79), Expect = 0.095, Method: Compositional matrix adjust.
Identities = 24/104 (23%), Positives = 49/104 (47%), Gaps = 4/104 (3%)
Query 18 QFKEVGRRAVRLW---LRDRSTLMAIMGDSVVQGLVIGAVFFGVR-SRASLYYQLSALFL 73
+ E+ A R W R R L+ +++V GLV+G ++ + +A + +
Sbjct 309 RITEISLLARRFWKIIYRTRQLLLTNALEALVVGLVLGTIYINIGIGKAGIEKRFGMFAF 368
Query 74 MILSLLASSLWTVPLYVQQKAQYRVEVEDGYYSPVPYMFATSLV 117
+ LL+S+ T+P+++ ++ E G Y ++ A +LV
Sbjct 369 TLTFLLSSTTETLPIFINERPILLRETSSGIYRLSSHILANTLV 412
> At5g19410
Length=624
Score = 32.3 bits (72), Expect = 0.64, Method: Compositional matrix adjust.
Identities = 24/93 (25%), Positives = 46/93 (49%), Gaps = 1/93 (1%)
Query 25 RAVRLWLRDRSTLMAIMGDSVVQGLVIGAVFFGVRSRASLYYQLSALFLMILS-LLASSL 83
R ++ R + +A +VV GL +G+V+ ++ + LF LS LL+S++
Sbjct 358 RFCKIIYRTKQLFLARTMQAVVAGLGLGSVYTRLKRDEEGVAERLGLFAFSLSFLLSSTV 417
Query 84 WTVPLYVQQKAQYRVEVEDGYYSPVPYMFATSL 116
+P+Y++++ E G Y YM A ++
Sbjct 418 EALPIYLRERRVLMKESSRGSYRISSYMIANTI 450
> At1g53270
Length=590
Score = 31.6 bits (70), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 26/121 (21%), Positives = 59/121 (48%), Gaps = 2/121 (1%)
Query 22 VGRRAVRLWLRDRSTLMAIMGDSVVQGLVIGAVFFGVRSRASLYYQL-SALFLMILS-LL 79
+G+R+ + R + + + GL++G+++ V ++ L + F IL+ LL
Sbjct 337 LGQRSCKNIFRTKQLFTTRALQASIAGLILGSIYLNVGNQKKEAKVLRTGFFAFILTFLL 396
Query 80 ASSLWTVPLYVQQKAQYRVEVEDGYYSPVPYMFATSLVANFFVLVGDAVLVTIMWLLFGF 139
+S+ +P+++Q + E Y + Y+ A +L+ F+L+ + T ++ L G
Sbjct 397 SSTTEGLPIFLQDRRILMRETSRRAYRVLSYVLADTLIFIPFLLIISMLFATPVYWLVGL 456
Query 140 Q 140
+
Sbjct 457 R 457
> CE05669
Length=695
Score = 30.8 bits (68), Expect = 1.6, Method: Composition-based stats.
Identities = 39/186 (20%), Positives = 78/186 (41%), Gaps = 31/186 (16%)
Query 24 RRAVRLWLRDRSTLMAIMGDSVVQGLVIGAVFFGVR-SRASL-------YYQLSALFLMI 75
R +++LW R+RS L+ + +++ ++IG+ ++G+ + SL + + + ++
Sbjct 420 RSSIQLW-RERSVLLVKLIQTLIMSIMIGSTYYGLEIDKKSLPSFKGFAFVSVQMMHMLF 478
Query 76 LSLLASSLWT-VPLYVQQKAQYRVEVEDGYYSPVPYMFATSLVANFFVLVGDAVLVTIMW 134
+ + W P+ V+ E + YSP Y A + + LV + I
Sbjct 479 MMPAMTVFWKDYPVVVR-------EFQANMYSPSAYYLAKTTADSIQYLVFPVIFSGI-- 529
Query 135 LLFGFQFVPLLACYLVGSLGFVICDLVTAI--CSLASKSFAEANASATLMFTL------L 186
L G +P + ++I +++ ++ CS+ A AT M L L
Sbjct 530 -LLGMTSLPYSVVIIT---NYLIINILLSLNACSVGQSFAAMCGHLATGMTVLPIVCVPL 585
Query 187 MFVNGF 192
M GF
Sbjct 586 MVFGGF 591
> At2g29940
Length=1443
Score = 30.4 bits (67), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 25/102 (24%), Positives = 48/102 (47%), Gaps = 5/102 (4%)
Query 44 SVVQGLVIGAVFFGVRSRAS----LYYQLSALFLMILSLLASSLWTV-PLYVQQKAQYRV 98
+ + ++G VF+ + S+ + L + AL+ L L S+ +V P+ ++ +
Sbjct 1195 TTIAAFILGTVFWDIGSKRTSSQDLITVMGALYSACLFLGVSNASSVQPIVSIERTVFYR 1254
Query 99 EVEDGYYSPVPYMFATSLVANFFVLVGDAVLVTIMWLLFGFQ 140
E G Y+P+PY A LV ++L + I + GF+
Sbjct 1255 EKAAGMYAPIPYAAAQGLVEIPYILTQTILYGVITYFTIGFE 1296
> At2g37280
Length=1413
Score = 28.9 bits (63), Expect = 7.1, Method: Composition-based stats.
Identities = 27/129 (20%), Positives = 59/129 (45%), Gaps = 9/129 (6%)
Query 18 QFKE-VGRRAVRLWLRDRSTLMAIMGDSVVQGLVIGAVFFG----VRSRASLYYQLSALF 72
QFK + + ++ W LM I G + + + G +F+ + ++ +L+ L A++
Sbjct 1139 QFKSCLWKMSLSYWRSPSYNLMRI-GHTFISSFIFGLLFWNQGKKIDTQQNLFTVLGAIY 1197
Query 73 LMILSLLASSLWTVPLY--VQQKAQYRVEVEDGYYSPVPYMFATSLVANFFVLVGDAVLV 130
++L + ++ + Y ++ YR E G YS Y A + ++ + A V
Sbjct 1198 GLVLFVGINNCTSALQYFETERNVMYR-ERFAGMYSAFAYALAQVVTEIPYIFIQSAEFV 1256
Query 131 TIMWLLFGF 139
+++ + GF
Sbjct 1257 IVIYPMIGF 1265
> At1g15520
Length=1423
Score = 28.5 bits (62), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 24/99 (24%), Positives = 49/99 (49%), Gaps = 5/99 (5%)
Query 49 LVIGAVFFGV----RSRASLYYQLSALFLMILSL-LASSLWTVPLYVQQKAQYRVEVEDG 103
L+ G +F+ + ++R L + +++ +L L L ++ P+ ++ + E G
Sbjct 1182 LMFGTMFWDLGGKTKTRQDLSNAMGSMYTAVLFLGLQNAASVQPVVNVERTVFYREQAAG 1241
Query 104 YYSPVPYMFATSLVANFFVLVGDAVLVTIMWLLFGFQFV 142
YS +PY FA + +VLV V I++ + GF++
Sbjct 1242 MYSAMPYAFAQVFIEIPYVLVQAIVYGLIVYAMIGFEWT 1280
Lambda K H
0.330 0.140 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3238438140
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40