bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2412_orf5
Length=324
Score E
Sequences producing significant alignments: (Bits) Value
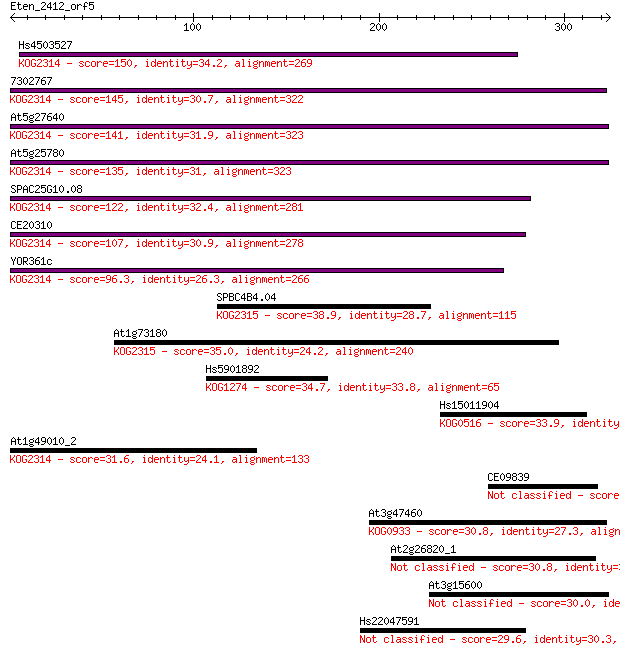
Hs4503527 150 4e-36
7302767 145 9e-35
At5g27640 141 2e-33
At5g25780 135 1e-31
SPAC25G10.08 122 1e-27
CE20310 107 4e-23
YOR361c 96.3 9e-20
SPBC4B4.04 38.9 0.014
At1g73180 35.0 0.22
Hs5901892 34.7 0.27
Hs15011904 33.9 0.46
At1g49010_2 31.6 2.3
CE09839 31.2 3.3
At3g47460 30.8 4.0
At2g26820_1 30.8 4.6
At3g15600 30.0 7.0
Hs22047591 29.6 9.2
> Hs4503527
Length=873
Score = 150 bits (378), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 92/270 (34%), Positives = 144/270 (53%), Gaps = 28/270 (10%)
Query 6 MHWQGNGDFLCLRTMVFKKTGKKGRKEFSQLEIFRMREKDIPVDNLQLNDIAVQLHWEEG 65
+HWQ NGD+LC++ +T K + + EIFRMREK +PVD +++ + + WE
Sbjct 517 LHWQKNGDYLCVKV---DRTPKGTQGVVTNFEIFRMREKQVPVDVVEMKETIIAFAWEPN 573
Query 66 PSKRFVLVVQEEGTTNQSLRFFRVADAAEGNVETNKRDTIMTHSFDIPNYMNFLKWSPFG 125
SK VL + S+ F+ V + G +E + FD N + WSP G
Sbjct 574 GSKFAVLHGE---APRISVSFYHVKN--NGKIE-------LIKMFD-KQQANTIFWSPQG 620
Query 126 SYFVLASL-STYGTVMFCHLNDQDKVEVLHQDEHYMCNEVRWSHCGRYLATCVVMPLVSG 184
+ VLA L S G + F +D V++ EHYM ++V W GRY+ T V
Sbjct 621 QFVVLAGLRSMNGALAFVDTSD---CTVMNIAEHYMASDVEWDPTGRYVVTSVSW----- 672
Query 185 AQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWRPHPPTLLTEKQLDDIKKRMKEHSK 244
+ + + +WTFQG+LL+K K+ F Q LWRP PPTLL+++Q+ IKK +K++SK
Sbjct 673 ---WSHKVDNAYWLWTFQGRLLQKNNKDRFCQLLWRPRPPTLLSQEQIKQIKKDLKKYSK 729
Query 245 RYEAEDERLRSEKRRAFLKQREEESEKFQQ 274
+E +D +S+ + +++R E F++
Sbjct 730 IFEQKDRLSQSKASKELVERRRTMMEDFRK 759
> 7302767
Length=690
Score = 145 bits (367), Expect = 9e-35, Method: Compositional matrix adjust.
Identities = 99/329 (30%), Positives = 169/329 (51%), Gaps = 33/329 (10%)
Query 1 VKGAAMHWQGNGDFLCLRTMVFKKTGKKGRKE-------FSQLEIFRMREKDIPVDNLQL 53
V +HWQ +GD+LC++ + K K + F EIF MREK+IPVD++++
Sbjct 384 VADCKLHWQKSGDYLCVKVDRYSKLKKDKKDLDVKFLGMFYNFEIFHMREKEIPVDSVEI 443
Query 54 NDIAVQLHWEEGPSKRFVLVVQEEGTTNQSLRFFRVADAAEGNVETNKRDTIMTHSFDIP 113
++ + WE +K F ++ E ++N S F+ V + ++ TH F
Sbjct 444 RELILAFAWEPVGNK-FSIIHGETNSSNVS--FYEVNKGVKPSLVKRLEKKSCTHLF--- 497
Query 114 NYMNFLKWSPFGSYFVLASLSTYGTVMFCHLNDQDKVEVLHQDEHYMCNEVRWSHCGRYL 173
WSP G + V+A+L T GT F ++ + + +H+ +EV W GRY+
Sbjct 498 -------WSPRGQFIVMANL-TMGTFEF--VDSTNDYIITASPDHFRASEVEWDPTGRYV 547
Query 174 ATCVVMPLVSGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWRPHPPTLLTEKQLD 233
V+G +++ ++TGFN++TFQG+++++T +NF QFLWRP PPTLL+E++
Sbjct 548 --------VTGVSSWKVKEDTGFNMYTFQGRIIKRTILKNFVQFLWRPRPPTLLSEEKQK 599
Query 234 DIKKRMKEHSKRYEAEDERLRSEKRRAFLKQREEESEKFQQILDELEAWKMKHPKYEEWC 293
+IKK +K++ +E +D + + L++R + E F + ++ A +
Sbjct 600 EIKKNLKKYYAAFEQKDRLRLTRASKELLEKRSQLRETFMEYRNKRIAEWAEQKSRRIML 659
Query 294 RAQEEFDTWFEWEYKEEIIEEELDVKQEV 322
R + D E EEI+ E VK+EV
Sbjct 660 RGHVDTDNLETDEVDEEIV--EFLVKEEV 686
> At5g27640
Length=738
Score = 141 bits (355), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 103/333 (30%), Positives = 171/333 (51%), Gaps = 40/333 (12%)
Query 1 VKGAAMHWQGNGDFLCLRTMVFKKTGKKGRKEFSQLEIFRMREKDIPVDNLQL---NDIA 57
V M+WQ +G++L ++ + KT K +S E+FR++E+DIP++ L+L ND
Sbjct 433 VSDCKMYWQSSGEYLAVKVDRYTKTKKS---TYSGFELFRIKERDIPIEVLELDNKNDKI 489
Query 58 VQLHWEEGPSKRFVLVVQEEGTTNQSLRFFRVADAAEGNVETNKRDTIMTHSFDIPNYMN 117
+ WE RF ++ ++ + S F A+ +K T+ N
Sbjct 490 IAFAWEPK-GHRFAVIHGDQPRPDVS---FYSMKTAQNTGRVSKLATLKAKQ------AN 539
Query 118 FLKWSPFGSYFVLASLSTY-GTVMFCHLNDQDKVEVLHQDEHYMCNEVRWSHCGRYLATC 176
L WSP G Y +LA L + G + F ++ D++E + EH+M ++ W GRY+AT
Sbjct 540 ALFWSPTGKYIILAGLKGFNGQLEFFNV---DELETMATAEHFMATDIEWDPTGRYVATA 596
Query 177 VVMPLVSGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWRPHPPTLLTEKQLDDIK 236
V + GF IW+F G +L + K++F+Q WRP PP+ LT ++ ++I
Sbjct 597 V-------TSVHEM--ENGFTIWSFNGIMLYRILKDHFFQLAWRPRPPSFLTAEKEEEIA 647
Query 237 KRMKEHSKRYEAEDER---LRSEKRRAFLKQREEESEKFQ---QILDELEAWKMKHPKYE 290
K +K++SK+YEAED+ L SE+ R K +EE EK+ + L E E ++ +
Sbjct 648 KTLKKYSKKYEAEDQDVSLLLSEQDREKRKALKEEWEKWVMQWKSLHEEEKLVRQNLRDG 707
Query 291 EWCRAQEEFDTWFEWEYKEEIIEEELDVKQEVI 323
E +E+ E+E KE E+ +DV +E++
Sbjct 708 EVSDVEED-----EYEAKEVEFEDLIDVTEEIV 735
> At5g25780
Length=730
Score = 135 bits (339), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 100/335 (29%), Positives = 166/335 (49%), Gaps = 44/335 (13%)
Query 1 VKGAAMHWQGNGDFLCLRTMVFKKTGKKGRKEFSQLEIFRMREKDIPVDNLQL---NDIA 57
V M+WQ +G++L ++ + KT K +S E+FR++E+DIP++ L+L ND
Sbjct 424 VSDCKMYWQSSGEYLAVKVDRYTKTKKS---TYSGFELFRIKERDIPIEVLELDNKNDKI 480
Query 58 VQLHWEEGPSKRFVLVVQEEGTTNQSLRFFRVADAAEGNVETNKRDTIMTHSFDIPNYMN 117
+ WE RF ++ ++ + S F A+ +K T+ N
Sbjct 481 IAFAWEP-KGHRFAVIHGDQPRPDVS---FYSMKTAQNTGRVSKLATLKAKQ------AN 530
Query 118 FLKWSPFGSYFVLASLSTY-GTVMFCHLNDQDKVEVLHQDEHYMCNEVRWSHCGRYLATC 176
L WSP G Y +LA L + G + F ++ D++E + EH+M ++ W GRY+AT
Sbjct 531 ALFWSPTGKYIILAGLKGFNGQLEFFNV---DELETMATAEHFMATDIEWDPTGRYVATS 587
Query 177 VVMPLVSGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWRPHPPTLLTEKQLDDIK 236
V V + GF IW+F G ++ + K++F+Q WRP P + LT ++ ++I
Sbjct 588 VTT--VHEME-------NGFTIWSFNGNMVYRILKDHFFQLAWRPRPASFLTAEKEEEIA 638
Query 237 KRMKEHSKRYEAEDERLRSEKRRAFLKQREEESEKFQQILDELEAWKM--KHPKYEEWCR 294
K ++ +SKRYEAED+ + L E++ EK + + +E + W M K EE
Sbjct 639 KNLRNYSKRYEAEDQDVS-------LLLSEQDREKRRALNEEWQKWVMQWKSLHEEEKLV 691
Query 295 AQEEFDTWF------EWEYKEEIIEEELDVKQEVI 323
Q D E+E KE E+ +DV +E++
Sbjct 692 RQNLRDGEVSDVEEDEYEAKEVEFEDLIDVTEEIV 726
> SPAC25G10.08
Length=725
Score = 122 bits (305), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 91/304 (29%), Positives = 148/304 (48%), Gaps = 48/304 (15%)
Query 1 VKGAAMHWQGNGDFLCLRTMVFKKTGKKGRKEFSQLEIFRMREKDIPVDNLQLNDIAVQL 60
V ++WQ NGD+LC++ KT K FS LEIFR+REK+IPV+ + L D+ +
Sbjct 406 VSDCKLYWQSNGDYLCVKVDRHTKTKKS---TFSNLEIFRIREKNIPVEVVDLKDVVLNF 462
Query 61 HWEEGPSKRFVLVVQEEGTTNQSLRFFRVADAAEGNVETN--------KRDTIMTH---- 108
WE S RF ++ + N + NV+TN K++T T
Sbjct 463 AWEPK-SDRFAIISANDQVLNST------------NVKTNLSFYGFEQKKNTPSTFRHII 509
Query 109 SFDIPNYMNFLKWSPFGSYFVLASL--STYGTVMFCHLN-DQDK--------VEVLHQDE 157
+FD N L +P G + V A+L ST + F L+ D +K V+ + E
Sbjct 510 TFD-KKTCNSLFMAPKGRFMVAATLGSSTQYDLEFYDLDFDTEKKEPDALANVQQIGSAE 568
Query 158 HYMCNEVRWSHCGRYLATCVVMPLVSGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQF 217
H+ E+ W GRY+ T + +R G+ + F+G LL + F QF
Sbjct 569 HFGMTELEWDPSGRYVTTSSTI--------WRHKLENGYRLCDFRGTLLREEMIGEFKQF 620
Query 218 LWRPHPPTLLTEKQLDDIKKRMKEHSKRYEAEDERLRSEKRRAFLKQREEESEKFQQILD 277
+WRP PP+ LT++ + I+K++K++++ ++ ED +S R +R + ++Q+ D
Sbjct 621 IWRPRPPSPLTKEDMKKIRKKLKDYNRLFDEEDIAEQSSANRELAARRRQLISEWQKYRD 680
Query 278 ELEA 281
E+ A
Sbjct 681 EVIA 684
> CE20310
Length=725
Score = 107 bits (267), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 86/299 (28%), Positives = 135/299 (45%), Gaps = 42/299 (14%)
Query 1 VKGAAMHWQGNGDFLCLRTMVFKKTGKKGRKEFS-------QLEIFRMREKDIPVDNLQL 53
V A M WQ +G L TM FKK + E ++IF + +KD+ + NL L
Sbjct 404 VADAQMFWQKSGKRLAFYTMRFKKKEYRETGEVKYVGGCQYHVDIFEIDKKDVSLMNLPL 463
Query 54 NDIAVQLHWEEGPSKRFVLVVQEEGTTNQSLRFFRVADAAEGNVETNKRDTIMTHSFDIP 113
++ + W+ K VLV T Q + +E N + D
Sbjct 464 SEPFIHFDWDPEGDKFCVLVGNTAKATPQVYK-----------IEANSHAPKLVSKLDAG 512
Query 114 NYMNFLKWSPFGSYF-VLASLSTYGTVMF--CHLNDQDKVEVLHQDEHYMCNEVRWSHCG 170
+ N ++++P G + VLA +S G V F L++ + V+ EH + N+ W G
Sbjct 513 VHFNEVQFAPKGGWLAVLAKVSAGGNVYFIDTSLSEAKRTNVI---EHPLFNKGYWDPTG 569
Query 171 RYLATCVVMPLVSGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWRPHPPTLLTEK 230
RY TC + R G + G+ I+TFQG+ L + + QF WRP PP L+E+
Sbjct 570 RYFVTCSTL-------GGRAGADLGYRIFTFQGRELCRKNLDRLAQFKWRPRPPVKLSEQ 622
Query 231 QLDDIKKRMKEHSKRY--EAEDERLRS-----EKRR----AFLKQREEESEKFQQILDE 278
+ +IKK +K+ + ++ + +DE+ R+ EKRR AF R E+ DE
Sbjct 623 KQREIKKNLKKTAAKFIKQDDDEKCRASQEVVEKRRKIMAAFDIIRSRNREQLDATRDE 681
> YOR361c
Length=763
Score = 96.3 bits (238), Expect = 9e-20, Method: Compositional matrix adjust.
Identities = 70/289 (24%), Positives = 129/289 (44%), Gaps = 39/289 (13%)
Query 1 VKGAAMHWQGNGDFLCLRTMVFKKTGKKGRKEFSQLEIFRMREKDIPVDNLQLNDIAVQL 60
V +HWQ +FLC ++ K G+ +FS L+I R+ E+DIPV+ ++L D +
Sbjct 436 VSNVTLHWQNQAEFLCFNV---ERHTKSGKTQFSNLQICRLTERDIPVEKVELKDSVFEF 492
Query 61 HWEEGPSKRFVLVVQEEGTTN-----QSLRFFRVADAAEGNVETNKRDTIMTHSFDIP-N 114
WE ++ + V E N ++RF+ A + + KR +++ +IP
Sbjct 493 GWEPHGNRFVTISVHEVADMNYAIPANTIRFY--APETKEKTDVIKRWSLVK---EIPKT 547
Query 115 YMNFLKWSPFGSYFVLASLSTYGT----VMFC--------HLNDQDKVEVLHQD----EH 158
+ N + WSP G + V+ +L + F ++ND + V +D +
Sbjct 548 FANTVSWSPAGRFVVVGALVGPNMRRSDLQFYDMDYPGEKNINDNNDVSASLKDVAHPTY 607
Query 159 YMCNEVRWSHCGRYLATCVVMPLVSGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFL 218
+ W GRY+ + + + + G+ I+ G L+++ F F
Sbjct 608 SAATNITWDPSGRYVT--------AWSSSLKHKVEHGYKIFNIAGNLVKEDIIAGFKNFA 659
Query 219 WRPHPPTLLTEKQLDDIKKRMKEHSKRYEAEDE-RLRSEKRRAFLKQRE 266
WRP P ++L+ + ++K ++E S ++E +D + R L QRE
Sbjct 660 WRPRPASILSNAERKKVRKNLREWSAQFEEQDAMEADTAMRDLILHQRE 708
> SPBC4B4.04
Length=576
Score = 38.9 bits (89), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 33/119 (27%), Positives = 47/119 (39%), Gaps = 15/119 (12%)
Query 113 PNYMNFLKWSPFGSYFVLASLSTY-GTV-MFCHLNDQDKVEVLHQDEHYMCNEVRWSHCG 170
P N L +SP Y +LA G++ +F N+ K+ + E C +S
Sbjct 309 PAPRNTLIFSPNSRYILLAGFGNLQGSIDIFDAANNMKKITTV---EAANCTYCEFSPDS 365
Query 171 RYLATCVVMPLVSGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWRPHP--PTLL 227
++L T V P R + IW G + + YQ WRP P PTLL
Sbjct 366 QFLLTAVTSP--------RLRVDNSIKIWHITGAPMFYEEFNELYQAFWRPRPLNPTLL 416
> At1g73180
Length=513
Score = 35.0 bits (79), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 58/280 (20%), Positives = 101/280 (36%), Gaps = 66/280 (23%)
Query 57 AVQLHWEEGPSKRFVLVVQEEGTTNQS------LRFFRVADAAEGNVETNKRDTI----- 105
+VQ W G + V+V + TNQS L + + EG V K +
Sbjct 221 SVQFSWNHGSTGLLVVVQSDVDKTNQSYYGETKLHYLTIDGTHEGLVPLRKEGPVHDVQW 280
Query 106 --------MTHSFDIPNYM------------------NFLKWSPFGSYFVLASLSTY-GT 138
+ + F +P + N L+W+P G +A G
Sbjct 281 SFSGSEFAVVYGF-MPACVTIFDKNCKPLMELGEGPYNTLRWNPKGRVLCVAGFGNLPGD 339
Query 139 VMFCHLNDQDKVEVLHQDEHYMCNEVRWSHCGRYLATCVVMPLVSGAQAYRFGDNTGFNI 198
+ F + ++ ++ ++ E + +E WS GRY T P R + G I
Sbjct 340 MAFWDVVNKKQLGS-NKAEWSVTSE--WSPDGRYFLTASTAP--------RRQIDNGMKI 388
Query 199 WTFQGKLLEKTKKENFYQFLWRPHPPTLLTEKQLDDIKKRMK--EHSKRYEAEDERLRSE 256
+ + GK K E YQ W+P P ++ DI + +K E K E + + S
Sbjct 389 FNYDGKRYYKKMFERLYQAEWKPESP-----ERFGDISELIKSVESLKLGEGKSQGQGSA 443
Query 257 KRRAFLKQREEESEKFQQILDELEAWKMKHPKYEEWCRAQ 296
+++ I + A++ H K+ +A+
Sbjct 444 QKKTIAP---------NPIAQKPAAYRPPHAKHAAAVQAE 474
> Hs5901892
Length=1129
Score = 34.7 bits (78), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 35/71 (49%), Gaps = 8/71 (11%)
Query 107 THSFD-----IPNYMNFLKWSPFGSYFVLASLSTYGTVMFCHLNDQDKVE-VLHQDEHYM 160
+H FD I +N + WSP G Y LA+ S G ++ ++ +D +E V H+ + +
Sbjct 219 SHQFDLSDNFISQTLNIVTWSPCGQY--LAAGSINGLIIVWNVETKDCMERVKHEKGYAI 276
Query 161 CNEVRWSHCGR 171
C CGR
Sbjct 277 CGLAWHPTCGR 287
> Hs15011904
Length=5430
Score = 33.9 bits (76), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 22/82 (26%), Positives = 42/82 (51%), Gaps = 5/82 (6%)
Query 233 DDIKKRMKEHSKRYEAEDERLRSEKRRAFLKQREEESEKFQQILDELEAWKMKHPKYEE- 291
+++ K M S + E + ++E +AFL + E+ S K Q++ + L + +P +E
Sbjct 2418 EEVLKSMDAMSSPTKTETVKAQAESNKAFLAELEQNSPKIQKVKEALAGLLVTYPNSQEA 2477
Query 292 --WCRAQEEFDTWFEWEYKEEI 311
W + QEE ++ WE E+
Sbjct 2478 ENWKKIQEELNS--RWERATEV 2497
> At1g49010_2
Length=241
Score = 31.6 bits (70), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 32/134 (23%), Positives = 54/134 (40%), Gaps = 20/134 (14%)
Query 1 VKGAAMHWQGNGDFLCLRTMVFKKTGKKGRKEFSQLEIFRMRE-KDIPVDNLQLNDIAVQ 59
V M+WQ G +L ++ R S E+FR + + I + L++++ +
Sbjct 107 VSDCKMYWQSKGKYLAVQVT---------RHTCSLFELFRFNKVQGITTEALEVDEKILA 157
Query 60 LHWEEGPSKRFVLVVQEEGTTNQSLRFFRVADAAEGNVETNKRDTIMTHSFDIPNYMNFL 119
WE RF ++ ++ N S F + + + +K T D L
Sbjct 158 FAWEPS-GHRFAVIHGDQSKPNVS---FYSMETPQNPGKVSKLATFDDKEADA------L 207
Query 120 KWSPFGSYFVLASL 133
WSP G + VLA L
Sbjct 208 FWSPRGKHIVLAGL 221
> CE09839
Length=318
Score = 31.2 bits (69), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 30/59 (50%), Gaps = 7/59 (11%)
Query 259 RAFLKQREEESEKFQQILDELEAWKMKHPKYEEWCRAQEEFDTWFEWEYKEEIIEEELD 317
+ L +E ++FQ+ + E E W + +Y+ WC A E+F W Y I+EE D
Sbjct 14 KVVLNPTQEYFDQFQEWITETERWNYRRTEYKLWCTAFEKF-----WLYMA--IDEEND 65
> At3g47460
Length=1171
Score = 30.8 bits (68), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 35/146 (23%), Positives = 62/146 (42%), Gaps = 18/146 (12%)
Query 195 GFNIWTFQGKLLEKTKKENFYQF--LWRPHPPTLLTEKQLDDI--KKRMKEHSKRYEAED 250
G N + GKL + + +N + L +P L+ + ++ + K M+ S EA
Sbjct 111 GKNKYLINGKLAQPNQVQNLFHSVQLNVNNPHFLIMQGRITKVLNMKPMEILSMLEEAAG 170
Query 251 ERLRSEKRRAFLKQREEESEKFQQI--------LDELEAWKMKHPKYEEWCRAQEEFD-- 300
R+ K+ A LK E++ K +I L LE + + +Y +W E D
Sbjct 171 TRMYENKKEAALKTLEKKQTKVDEINKLLEKDILPALEKLRREKSQYMQWANGNAELDRL 230
Query 301 ----TWFEWEYKEEIIEEELDVKQEV 322
FE+ E+I + + V +E+
Sbjct 231 KRFCVAFEYVQAEKIRDNSIHVVEEM 256
> At2g26820_1
Length=300
Score = 30.8 bits (68), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 35/112 (31%), Positives = 46/112 (41%), Gaps = 6/112 (5%)
Query 207 EKTKKENFYQFLWRPHPPTLLTEKQLDDIKKRMKEHSKRYEAEDERLRSEKRRAFLKQRE 266
EK K E Q L R +Q I + H K E DERLR E+R K R
Sbjct 176 EKKKVEQVKQLLARVEN----VGEQTGGIPYTYQLHRKIKEENDERLREEERVIESKNRA 231
Query 267 EE--SEKFQQILDELEAWKMKHPKYEEWCRAQEEFDTWFEWEYKEEIIEEEL 316
E +E Q +L E E +M+ K ++ E + E E + E EL
Sbjct 232 EAELAEMQQNLLMEKEKLQMEEAKNKQLIAQAEANEKLMEQERAKNRAETEL 283
> At3g15600
Length=591
Score = 30.0 bits (66), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 32/103 (31%), Positives = 50/103 (48%), Gaps = 15/103 (14%)
Query 227 LTEKQLDDIKKRMKEHSKRYEAEDERLRSEKRRAFLKQ-----REEESEKFQQILDELEA 281
L E + KRMK+ SK+YE +D+RL+ A +K R E++ ++I D
Sbjct 343 LMENGFKSMNKRMKDFSKKYEEQDKRLKL--LEAAIKSIQSGIRTEDACGSKEIDD---- 396
Query 282 WKMKHPKYEEWCRAQEEFDTWFEWEYKE-EIIEEELDVKQEVI 323
K + EE A+ E D KE E+ E E ++ +EV+
Sbjct 397 ---KENELEEGSDAETEIDKEVAQGDKEREVGETETEIDKEVV 436
> Hs22047591
Length=179
Score = 29.6 bits (65), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 49/99 (49%), Gaps = 11/99 (11%)
Query 190 FGDNTGFNIWTF---QGKLLEKTKKENFYQFLWRPHPPTLLTEKQLDDIKKR---MKEHS 243
+ + NI+T+ + K + K ++ + +P P + L KK+ ++E
Sbjct 26 YPEPRNINIYTYDDMEVKQINKRASGQAFELILKPPSPISEAPRTLASPKKKDLSLEEIQ 85
Query 244 KRYEAEDERLRSEKRRAFLKQ----REEESEKFQQILDE 278
K+ EA +ER +S++ + LKQ RE E E Q+ L+E
Sbjct 86 KKLEAAEERRKSQEAQ-VLKQLAEKREHEREVLQKALEE 123
Lambda K H
0.320 0.135 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7566324290
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40