bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2455_orf1
Length=178
Score E
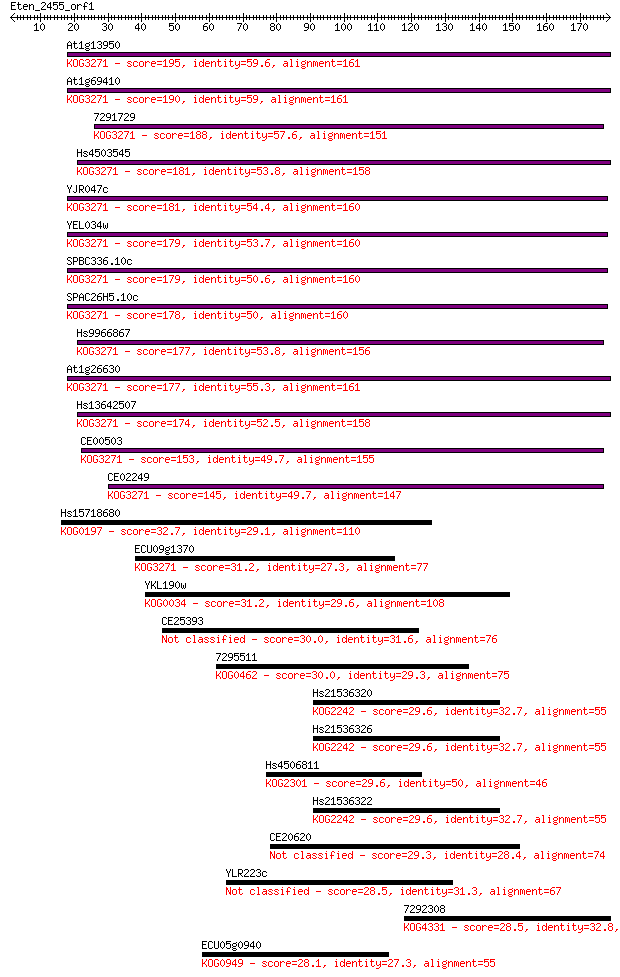
Sequences producing significant alignments: (Bits) Value
At1g13950 195 3e-50
At1g69410 190 2e-48
7291729 188 5e-48
Hs4503545 181 7e-46
YJR047c 181 1e-45
YEL034w 179 2e-45
SPBC336.10c 179 3e-45
SPAC26H5.10c 178 6e-45
Hs9966867 177 8e-45
At1g26630 177 9e-45
Hs13642507 174 6e-44
CE00503 153 2e-37
CE02249 145 5e-35
Hs15718680 32.7 0.36
ECU09g1370 31.2 1.0
YKL190w 31.2 1.1
CE25393 30.0 2.3
7295511 30.0 2.5
Hs21536320 29.6 3.0
Hs21536326 29.6 3.5
Hs4506811 29.6 3.6
Hs21536322 29.6 3.7
CE20620 29.3 4.5
YLR223c 28.5 7.5
7292308 28.5 7.6
ECU05g0940 28.1 8.9
> At1g13950
Length=158
Score = 195 bits (496), Expect = 3e-50, Method: Compositional matrix adjust.
Identities = 96/161 (59%), Positives = 123/161 (76%), Gaps = 6/161 (3%)
Query 18 MSDAEEVSFERADAGASLTYPQQAGAIKKNGFCMLKGNPCKVVDYSTSKTGKHGHAKAHI 77
MSD EE FE +DAGAS TYPQQAG I+KNG+ ++K PCKVV+ STSKTGKHGHAK H
Sbjct 1 MSD-EEHHFESSDAGASKTYPQQAGTIRKNGYIVIKNRPCKVVEVSTSKTGKHGHAKCHF 59
Query 78 VGLDIFTGKKYEDICPTSHNMEVPNVKRSEFQLIDVSDDGFVSLLLDNGDLKSDLTLPKD 137
V +DIFT KK EDI P+SHN +VP+V R+++QLID+S+DG+VSLL DNG K DL LP D
Sbjct 60 VAIDIFTSKKLEDIVPSSHNCDVPHVNRTDYQLIDISEDGYVSLLTDNGSTKDDLKLPND 119
Query 138 SDGNLDEVALQVKSLFQEGKGVLVSVLAACGKEKIVACKEL 178
D + Q+KS F +GK ++VSV++A G+E+I A K++
Sbjct 120 -----DTLLQQIKSGFDDGKDLVVSVMSAMGEEQINALKDI 155
> At1g69410
Length=158
Score = 190 bits (482), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 95/161 (59%), Positives = 122/161 (75%), Gaps = 6/161 (3%)
Query 18 MSDAEEVSFERADAGASLTYPQQAGAIKKNGFCMLKGNPCKVVDYSTSKTGKHGHAKAHI 77
MSD +E FE +DAGAS TYPQQAG I+K G ++KG PCKVV+ STSKTGKHGHAK H
Sbjct 1 MSD-DEHHFESSDAGASKTYPQQAGNIRKGGHIVIKGRPCKVVEVSTSKTGKHGHAKCHF 59
Query 78 VGLDIFTGKKYEDICPTSHNMEVPNVKRSEFQLIDVSDDGFVSLLLDNGDLKSDLTLPKD 137
V +DIFT KK EDI P+SHN +VP+V R ++QLID+S+DGFVSLL DNG K DL LP D
Sbjct 60 VAIDIFTSKKLEDIVPSSHNCDVPHVNRVDYQLIDISEDGFVSLLTDNGSTKDDLKLPTD 119
Query 138 SDGNLDEVALQVKSLFQEGKGVLVSVLAACGKEKIVACKEL 178
+ + Q+K+ F+EGK ++VSV++A G+E++ A KE+
Sbjct 120 -----EALLTQLKNGFEEGKDIVVSVMSAMGEEQMCALKEV 155
> 7291729
Length=159
Score = 188 bits (477), Expect = 5e-48, Method: Compositional matrix adjust.
Identities = 87/151 (57%), Positives = 115/151 (76%), Gaps = 6/151 (3%)
Query 26 FERADAGASLTYPQQAGAIKKNGFCMLKGNPCKVVDYSTSKTGKHGHAKAHIVGLDIFTG 85
FE D+GAS TYP Q A++KNGF MLK PCK+V+ STSKTGKHGHAK H+VG+DIF+
Sbjct 8 FETTDSGASTTYPMQCSALRKNGFVMLKSRPCKIVEMSTSKTGKHGHAKVHMVGIDIFSN 67
Query 86 KKYEDICPTSHNMEVPNVKRSEFQLIDVSDDGFVSLLLDNGDLKSDLTLPKDSDGNLDEV 145
KKYEDICP++HNM+VPNVKR + QLI +SDD F++L+ ++GDL+ DL +P +G L E
Sbjct 68 KKYEDICPSTHNMDVPNVKREDLQLIAISDDSFLTLMTESGDLREDLKVP---EGELGE- 123
Query 146 ALQVKSLFQEGKGVLVSVLAACGKEKIVACK 176
Q++ F GK +L +VL ACG+E ++A K
Sbjct 124 --QLRLDFDSGKDLLCTVLKACGEECVIAIK 152
> Hs4503545
Length=154
Score = 181 bits (459), Expect = 7e-46, Method: Compositional matrix adjust.
Identities = 85/158 (53%), Positives = 123/158 (77%), Gaps = 7/158 (4%)
Query 21 AEEVSFERADAGASLTYPQQAGAIKKNGFCMLKGNPCKVVDYSTSKTGKHGHAKAHIVGL 80
A+++ FE DAGAS T+P Q A++KNGF +LKG PCK+V+ STSKTGKHGHAK H+VG+
Sbjct 2 ADDLDFETGDAGASATFPMQCSALRKNGFVVLKGRPCKIVEMSTSKTGKHGHAKVHLVGI 61
Query 81 DIFTGKKYEDICPTSHNMEVPNVKRSEFQLIDVSDDGFVSLLLDNGDLKSDLTLPKDSDG 140
DIFTGKKYEDICP++HNM+VPN+KR++FQLI + DG++SLL D+G+++ DL LP +G
Sbjct 62 DIFTGKKYEDICPSTHNMDVPNIKRNDFQLIGIQ-DGYLSLLQDSGEVREDLRLP---EG 117
Query 141 NLDEVALQVKSLFQEGKGVLVSVLAACGKEKIVACKEL 178
+L + +++ + G+ +L++VL+A +E VA K +
Sbjct 118 DLGK---EIEQKYDCGEEILITVLSAMTEEAAVAIKAM 152
> YJR047c
Length=157
Score = 181 bits (458), Expect = 1e-45, Method: Compositional matrix adjust.
Identities = 87/160 (54%), Positives = 121/160 (75%), Gaps = 8/160 (5%)
Query 18 MSDAEEVSFERADAGASLTYPQQAGAIKKNGFCMLKGNPCKVVDYSTSKTGKHGHAKAHI 77
MSD EE +FE ADAGAS TYP Q A++KNGF ++KG PCK+VD STSKTGKHGHAK H+
Sbjct 1 MSD-EEHTFENADAGASATYPMQCSALRKNGFVVIKGRPCKIVDMSTSKTGKHGHAKVHL 59
Query 78 VGLDIFTGKKYEDICPTSHNMEVPNVKRSEFQLIDVSDDGFVSLLLDNGDLKSDLTLPKD 137
V LDIFTGKK ED+ P++HN+EVP VKRSE+QL+D+ DDG++SL+ +G+ K D+ P+
Sbjct 60 VTLDIFTGKKLEDLSPSTHNLEVPFVKRSEYQLLDI-DDGYLSLMTMDGETKDDVKAPEG 118
Query 138 SDGNLDEVALQVKSLFQEGKGVLVSVLAACGKEKIVACKE 177
G+ +++ F EGK ++V++++A G+E ++ KE
Sbjct 119 ELGD------SMQAAFDEGKDLMVTIISAMGEEAAISFKE 152
> YEL034w
Length=157
Score = 179 bits (454), Expect = 2e-45, Method: Compositional matrix adjust.
Identities = 86/160 (53%), Positives = 120/160 (75%), Gaps = 8/160 (5%)
Query 18 MSDAEEVSFERADAGASLTYPQQAGAIKKNGFCMLKGNPCKVVDYSTSKTGKHGHAKAHI 77
MSD EE +FE ADAG+S TYP Q A++KNGF ++K PCK+VD STSKTGKHGHAK H+
Sbjct 1 MSD-EEHTFETADAGSSATYPMQCSALRKNGFVVIKSRPCKIVDMSTSKTGKHGHAKVHL 59
Query 78 VGLDIFTGKKYEDICPTSHNMEVPNVKRSEFQLIDVSDDGFVSLLLDNGDLKSDLTLPKD 137
V +DIFTGKK ED+ P++HNMEVP VKR+E+QL+D+ DDGF+SL+ +GD K D+ P+
Sbjct 60 VAIDIFTGKKLEDLSPSTHNMEVPVVKRNEYQLLDI-DDGFLSLMNMDGDTKDDVKAPEG 118
Query 138 SDGNLDEVALQVKSLFQEGKGVLVSVLAACGKEKIVACKE 177
G+ +++ F EGK ++V++++A G+E ++ KE
Sbjct 119 ELGD------SLQTAFDEGKDLMVTIISAMGEEAAISFKE 152
> SPBC336.10c
Length=157
Score = 179 bits (453), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 81/160 (50%), Positives = 120/160 (75%), Gaps = 7/160 (4%)
Query 18 MSDAEEVSFERADAGASLTYPQQAGAIKKNGFCMLKGNPCKVVDYSTSKTGKHGHAKAHI 77
M++ E V FE +AGASLT+P Q A++KNG ++KG PCK+VD STSKTGKHGHAK HI
Sbjct 1 MAEEEHVDFEGGEAGASLTFPMQCSALRKNGHVVIKGRPCKIVDMSTSKTGKHGHAKVHI 60
Query 78 VGLDIFTGKKYEDICPTSHNMEVPNVKRSEFQLIDVSDDGFVSLLLDNGDLKSDLTLPKD 137
V LDIF G+KYED+ P++HNM+VP VKR E+QL+++ DDG+++L+ +G K D+ LP+
Sbjct 61 VALDIFNGRKYEDMSPSTHNMDVPVVKRDEYQLVNI-DDGYLNLMTTDGTTKDDVRLPEG 119
Query 138 SDGNLDEVALQVKSLFQEGKGVLVSVLAACGKEKIVACKE 177
GN +++ F+EGK ++++V++A G+E +AC++
Sbjct 120 ELGN------EIEEGFEEGKDLIITVVSAMGEEIALACRD 153
> SPAC26H5.10c
Length=157
Score = 178 bits (451), Expect = 6e-45, Method: Compositional matrix adjust.
Identities = 80/160 (50%), Positives = 119/160 (74%), Gaps = 7/160 (4%)
Query 18 MSDAEEVSFERADAGASLTYPQQAGAIKKNGFCMLKGNPCKVVDYSTSKTGKHGHAKAHI 77
M++ E V FE +AGASLT+P Q A++KNG ++KG PCK+VD STSKTGKHGHAK HI
Sbjct 1 MAEEEHVDFEGGEAGASLTFPMQCSALRKNGHVVIKGRPCKIVDMSTSKTGKHGHAKVHI 60
Query 78 VGLDIFTGKKYEDICPTSHNMEVPNVKRSEFQLIDVSDDGFVSLLLDNGDLKSDLTLPKD 137
V LDIF G+KYED+ P++HNM+VP VKR E+QL+++ DDG+++L+ +G K D+ LP+
Sbjct 61 VALDIFNGRKYEDMSPSTHNMDVPVVKRDEYQLVNI-DDGYLNLMTTDGTTKDDVRLPEG 119
Query 138 SDGNLDEVALQVKSLFQEGKGVLVSVLAACGKEKIVACKE 177
GN +++ F EG+ ++++V++A G+E +AC++
Sbjct 120 ELGN------EIEEGFDEGRDLIITVVSAMGEETALACRD 153
> Hs9966867
Length=153
Score = 177 bits (450), Expect = 8e-45, Method: Compositional matrix adjust.
Identities = 84/156 (53%), Positives = 116/156 (74%), Gaps = 7/156 (4%)
Query 21 AEEVSFERADAGASLTYPQQAGAIKKNGFCMLKGNPCKVVDYSTSKTGKHGHAKAHIVGL 80
A+E+ F DAGAS TYP Q A++KNGF +LKG PCK+V+ STSKTGKHGHAK H+VG+
Sbjct 2 ADEIDFTTGDAGASSTYPMQCSALRKNGFVVLKGRPCKIVEMSTSKTGKHGHAKVHLVGI 61
Query 81 DIFTGKKYEDICPTSHNMEVPNVKRSEFQLIDVSDDGFVSLLLDNGDLKSDLTLPKDSDG 140
DIFTGKKYEDICP++HNM+VPN+KR+++QLI + DG++SLL + G+++ DL LP+
Sbjct 62 DIFTGKKYEDICPSTHNMDVPNIKRNDYQLICIQ-DGYLSLLTETGEVREDLKLPE---- 116
Query 141 NLDEVALQVKSLFQEGKGVLVSVLAACGKEKIVACK 176
E+ +++ + G+ V VSV+ A +E VA K
Sbjct 117 --GELGKEIEGKYNAGEDVQVSVMCAMSEEYAVAIK 150
> At1g26630
Length=159
Score = 177 bits (449), Expect = 9e-45, Method: Compositional matrix adjust.
Identities = 89/161 (55%), Positives = 118/161 (73%), Gaps = 6/161 (3%)
Query 18 MSDAEEVSFERADAGASLTYPQQAGAIKKNGFCMLKGNPCKVVDYSTSKTGKHGHAKAHI 77
MSD +E FE +++GAS TYPQ AG I+K G ++K PCKVV+ STSKTGKHGHAK H
Sbjct 1 MSD-DEHHFEASESGASKTYPQSAGNIRKGGHIVIKNRPCKVVEVSTSKTGKHGHAKCHF 59
Query 78 VGLDIFTGKKYEDICPTSHNMEVPNVKRSEFQLIDVSDDGFVSLLLDNGDLKSDLTLPKD 137
V +DIFT KK EDI P+SHN +VP+V R ++QLID+++DGFVSLL D+G K DL LP D
Sbjct 60 VAIDIFTAKKLEDIVPSSHNCDVPHVNRVDYQLIDITEDGFVSLLTDSGGTKDDLKLPTD 119
Query 138 SDGNLDEVALQVKSLFQEGKGVLVSVLAACGKEKIVACKEL 178
D + Q++ F EGK ++VSV+++ G+E+I A KE+
Sbjct 120 -----DGLTAQMRLGFDEGKDIVVSVMSSMGEEQICAVKEV 155
> Hs13642507
Length=154
Score = 174 bits (442), Expect = 6e-44, Method: Compositional matrix adjust.
Identities = 83/158 (52%), Positives = 120/158 (75%), Gaps = 7/158 (4%)
Query 21 AEEVSFERADAGASLTYPQQAGAIKKNGFCMLKGNPCKVVDYSTSKTGKHGHAKAHIVGL 80
A+++ FE DAGAS T+P Q A++KNGF +LKG PCK+V+ S SKTGKHGHAK H+VG+
Sbjct 2 ADDLDFETGDAGASATFPMQCSALRKNGFVVLKGWPCKIVEMSASKTGKHGHAKVHLVGI 61
Query 81 DIFTGKKYEDICPTSHNMEVPNVKRSEFQLIDVSDDGFVSLLLDNGDLKSDLTLPKDSDG 140
DIFTGKKYEDICP++HNM+VPN++R++FQLI + DG++SLL D+G++ DL LP +G
Sbjct 62 DIFTGKKYEDICPSTHNMDVPNIRRNDFQLIGIQ-DGYLSLLQDSGEVPEDLRLP---EG 117
Query 141 NLDEVALQVKSLFQEGKGVLVSVLAACGKEKIVACKEL 178
+L + + + + G+ +L++VL+A +E VA K +
Sbjct 118 DLGK---ETEQKYDCGEEILITVLSAMTEEAAVAIKAM 152
> CE00503
Length=161
Score = 153 bits (386), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 77/156 (49%), Positives = 111/156 (71%), Gaps = 7/156 (4%)
Query 22 EEVSFERADAGASLTYPQQAGAIKKNGFCMLKGNPCKVVDYSTSKTGKHGHAKAHIVGLD 81
+E F+ A++GA+ T+P+Q A++KN M++G PCK+V+ STSKTGKHGHAK H+V +D
Sbjct 7 DEEQFDSAESGAAATFPKQCSALRKNEHVMIRGRPCKIVEMSTSKTGKHGHAKVHMVAID 66
Query 82 IFTGKKYEDICPTSHNMEVPNVKRSEFQLIDVSDDGFVSLL-LDNGDLKSDLTLPKDSDG 140
IFT KK EDICP++HNM+VP VKR E+ L+ + +DGF SL+ ++ +LK DL +P+ G
Sbjct 67 IFTTKKLEDICPSTHNMDVPVVKRREYILMSI-EDGFCSLMDPESCELKDDLKMPEGDLG 125
Query 141 NLDEVALQVKSLFQEGKGVLVSVLAACGKEKIVACK 176
N AL+ ++ VLV V+AACG+E I+ K
Sbjct 126 NTIREALE-----KDEGSVLVQVVAACGEEAILGYK 156
> CE02249
Length=161
Score = 145 bits (365), Expect = 5e-35, Method: Compositional matrix adjust.
Identities = 73/149 (48%), Positives = 106/149 (71%), Gaps = 9/149 (6%)
Query 30 DAGASLTYPQQAGAIKKNGFCMLKGNPCKVVDYSTSKTGKHGHAKAHIVGLDIFTGKKYE 89
D+GA+ T+P+Q A++KN M+KG PCK+V+ STSKTGKHGHAK H+V +DIFT KK E
Sbjct 15 DSGAAATFPKQCSALRKNEHVMIKGRPCKIVEMSTSKTGKHGHAKVHMVAIDIFTSKKLE 74
Query 90 DICPTSHNMEVPNVKRSEFQLIDVSDDGFVSLL-LDNGDLKSDLTLPKDSDGNLDEVALQ 148
DICP++HNM+VP VKR E+ L+ + DDG+ SL+ ++ + K DL LP E+ Q
Sbjct 75 DICPSTHNMDVPVVKRREYLLMAI-DDGYCSLMDPESCEQKDDLKLPD------TELGQQ 127
Query 149 VKSLFQEGKG-VLVSVLAACGKEKIVACK 176
++ +++ +G VLV V++A G+E I+ K
Sbjct 128 IRDAYEKDEGSVLVQVVSAIGEEAILGWK 156
> Hs15718680
Length=620
Score = 32.7 bits (73), Expect = 0.36, Method: Composition-based stats.
Identities = 32/120 (26%), Positives = 53/120 (44%), Gaps = 15/120 (12%)
Query 16 AAMSDAEEVSFERADAGASLTYPQQAGAIKKNGFCMLKGNPCKVVDYSTS-------KTG 68
AMS EE E A+ L++P+ ++ G C+ + C V ++ +T
Sbjct 396 GAMS--EEDFIEEAEVMMKLSHPK---LVQLYGVCLEQAPICLVFEFMEHGCLSDYLRTQ 450
Query 69 KHGHAKAHIVG--LDIFTGKKY-EDICPTSHNMEVPNVKRSEFQLIDVSDDGFVSLLLDN 125
+ A ++G LD+ G Y E+ C ++ N E Q+I VSD G +LD+
Sbjct 451 RGLFAAETLLGMCLDVCEGMAYLEEACVIHRDLAARNCLVGENQVIKVSDFGMTRFVLDD 510
> ECU09g1370
Length=161
Score = 31.2 bits (69), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 40/81 (49%), Gaps = 4/81 (4%)
Query 38 PQQAGAIKKNGFCMLKGN----PCKVVDYSTSKTGKHGHAKAHIVGLDIFTGKKYEDICP 93
P I+ + ++ N +V++ S+ K GKHG AK I + TG ++ I
Sbjct 10 PTSPKDIRSGNYIKIEVNGQMKSAQVLETSSVKNGKHGAAKTTISSKILSTGSNHKGIYT 69
Query 94 TSHNMEVPNVKRSEFQLIDVS 114
+ ++ V ++ + +LID+S
Sbjct 70 ANDSIIVCRPEKVQLKLIDIS 90
> YKL190w
Length=175
Score = 31.2 bits (69), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 32/113 (28%), Positives = 51/113 (45%), Gaps = 22/113 (19%)
Query 41 AGAIKKNGFCMLKG---NPC--KVVDYSTSKTGKHGHAKAHIVGLDIFTGKKYEDICPTS 95
+G+I KN F + G NP ++++ + + I GL IF+G+ +D
Sbjct 38 SGSIDKNEFMSIPGVSSNPLAGRIMEVFDADNSGDVDFQEFITGLSIFSGRGSKD----- 92
Query 96 HNMEVPNVKRSEFQLIDVSDDGFVSLLLDNGDLKSDLTLPKDSDGNLDEVALQ 148
R F++ D+ DGF+S NG+L L + S NLD+ LQ
Sbjct 93 ------EKLRFAFKIYDIDKDGFIS----NGELFIVLKIMVGS--NLDDEQLQ 133
> CE25393
Length=315
Score = 30.0 bits (66), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 34/88 (38%), Gaps = 12/88 (13%)
Query 46 KNGF------------CMLKGNPCKVVDYSTSKTGKHGHAKAHIVGLDIFTGKKYEDICP 93
KNGF LK K+V+Y + + V L I ++ +
Sbjct 21 KNGFFFSLSCGYAAFPLFLKVFSLKIVNYWRERQNDERKQQKEFVDLPIHVVQRIMEKVE 80
Query 94 TSHNMEVPNVKRSEFQLIDVSDDGFVSL 121
S + V V R+ QLID D GF S+
Sbjct 81 PSDRLSVRKVNRNLRQLIDEMDPGFRSI 108
> 7295511
Length=679
Score = 30.0 bits (66), Expect = 2.5, Method: Composition-based stats.
Identities = 22/85 (25%), Positives = 36/85 (42%), Gaps = 10/85 (11%)
Query 62 YSTSKTGKHGHAKAHIVGLDI-FTGKKYEDICPTSHNMEVPNVKRSEF---------QLI 111
Y++ HG+ +H+V LDI GK E++C H + V R Q++
Sbjct 567 YASFSYEDHGYHPSHLVRLDIHLNGKPVEELCRIVHVSKATGVARQMVLKLRELIPKQMV 626
Query 112 DVSDDGFVSLLLDNGDLKSDLTLPK 136
++ V L GD+ + L K
Sbjct 627 QIAIQACVGSKLYGGDVTRRMKLLK 651
> Hs21536320
Length=756
Score = 29.6 bits (65), Expect = 3.0, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 30/55 (54%), Gaps = 8/55 (14%)
Query 91 ICPTSHNMEVPNVKRSEFQLIDVSDDGFVSLLLDNGDLKSDLTLPKDSDGNLDEV 145
ICPT +++ +KR++ + DV D + ++K++ TLP D LDEV
Sbjct 431 ICPTDEDLKDRTIKRTDEEGKDVPDHAVL-------EMKANFTLPDVGD-FLDEV 477
> Hs21536326
Length=856
Score = 29.6 bits (65), Expect = 3.5, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 30/55 (54%), Gaps = 8/55 (14%)
Query 91 ICPTSHNMEVPNVKRSEFQLIDVSDDGFVSLLLDNGDLKSDLTLPKDSDGNLDEV 145
ICPT +++ +KR++ + DV D + ++K++ TLP D LDEV
Sbjct 531 ICPTDEDLKDRTIKRTDEEGKDVPDHAVL-------EMKANFTLPDVGD-FLDEV 577
> Hs4506811
Length=1682
Score = 29.6 bits (65), Expect = 3.6, Method: Composition-based stats.
Identities = 23/63 (36%), Positives = 31/63 (49%), Gaps = 17/63 (26%)
Query 77 IVGLDIFTGKKYEDICPTSH----NMEVPNVKRSEFQLIDVS-------------DDGFV 119
I+G+D+F G+ YE I PTS + EV N R E L + S +GF+
Sbjct 1074 IMGVDLFAGRFYECIDPTSGERFPSSEVMNKSRCESLLFNESMLWENAKMNFDNVGNGFL 1133
Query 120 SLL 122
SLL
Sbjct 1134 SLL 1136
> Hs21536322
Length=804
Score = 29.6 bits (65), Expect = 3.7, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 30/55 (54%), Gaps = 8/55 (14%)
Query 91 ICPTSHNMEVPNVKRSEFQLIDVSDDGFVSLLLDNGDLKSDLTLPKDSDGNLDEV 145
ICPT +++ +KR++ + DV D + ++K++ TLP D LDEV
Sbjct 531 ICPTDEDLKDRTIKRTDEEGKDVPDHAVL-------EMKANFTLPDVGD-FLDEV 577
> CE20620
Length=786
Score = 29.3 bits (64), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 34/74 (45%), Gaps = 4/74 (5%)
Query 78 VGLDIFTGKKYEDICPTSHNMEVPNVKRSEFQLIDVSDDGFVSLLLDNGDLKSDLTLPKD 137
GL + T + + P H VK+ +F+ + G L+L+ + L KD
Sbjct 604 TGLRLLTADELQPQNPKYHAW----VKKDQFRNAAPATGGIGRLMLEKMGWRPGEGLGKD 659
Query 138 SDGNLDEVALQVKS 151
+ GNL+ + L VKS
Sbjct 660 ATGNLEPLMLDVKS 673
> YLR223c
Length=1085
Score = 28.5 bits (62), Expect = 7.5, Method: Composition-based stats.
Identities = 21/69 (30%), Positives = 36/69 (52%), Gaps = 8/69 (11%)
Query 65 SKTGKHGHAKAHIVGLDIFTG--KKYEDICPTSHNMEVPNVKRSEFQLIDVSDDGFVSLL 122
SK G+H K+HI + + K +D+ P++H+ + + K D SDD + ++L
Sbjct 515 SKKGRHKSGKSHIEHKNKGSNLIKSNDDLEPSTHSTVLNSGK------YDSSDDEYDNIL 568
Query 123 LDNGDLKSD 131
LD + SD
Sbjct 569 LDVAHMPSD 577
> 7292308
Length=886
Score = 28.5 bits (62), Expect = 7.6, Method: Composition-based stats.
Identities = 20/61 (32%), Positives = 29/61 (47%), Gaps = 1/61 (1%)
Query 118 FVSLLLDNGDLKSDLTLPKDSDGNLDEVALQVKSLFQEGKGVLVSVLAACGKEKIVACKE 177
++ LLD DL L +DS DEVA + L + G V V+ KE + +C+
Sbjct 454 YLHHLLDKIKQGDDL-LRQDSKKEADEVARNISKLVKSGLDDYVRVVEEATKEDMESCEP 512
Query 178 L 178
L
Sbjct 513 L 513
> ECU05g0940
Length=1337
Score = 28.1 bits (61), Expect = 8.9, Method: Composition-based stats.
Identities = 15/55 (27%), Positives = 29/55 (52%), Gaps = 0/55 (0%)
Query 58 KVVDYSTSKTGKHGHAKAHIVGLDIFTGKKYEDICPTSHNMEVPNVKRSEFQLID 112
++ DYS KH ++ VG+++ ++I PTS + + N+++S L D
Sbjct 257 ELTDYSDVVGDKHRDIESRNVGMELDLLVSGKEISPTSAAVHMKNIRKSAQSLFD 311
Lambda K H
0.315 0.131 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2779358582
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40