bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
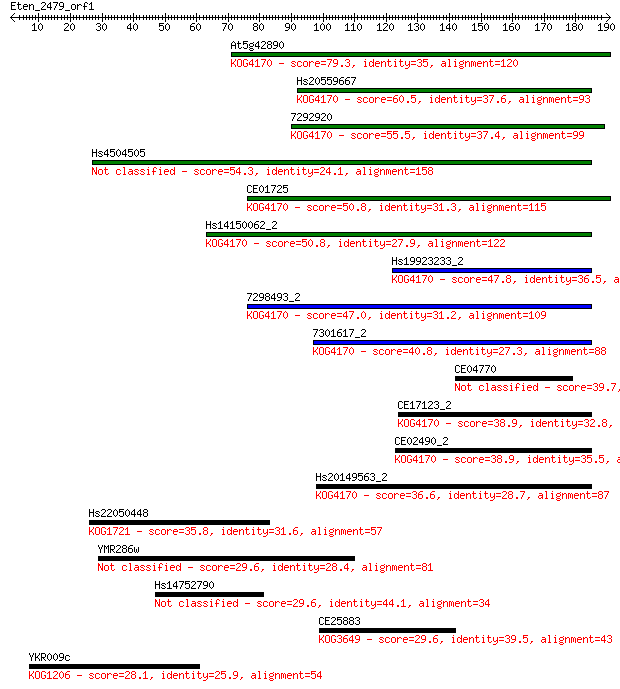
Query= Eten_2479_orf1
Length=190
Score E
Sequences producing significant alignments: (Bits) Value
At5g42890 79.3 4e-15
Hs20559667 60.5 2e-09
7292920 55.5 6e-08
Hs4504505 54.3 2e-07
CE01725 50.8 2e-06
Hs14150062_2 50.8 2e-06
Hs19923233_2 47.8 1e-05
7298493_2 47.0 2e-05
7301617_2 40.8 0.002
CE04770 39.7 0.004
CE17123_2 38.9 0.005
CE02490_2 38.9 0.006
Hs20149563_2 36.6 0.035
Hs22050448 35.8 0.051
YMR286w 29.6 3.5
Hs14752790 29.6 3.8
CE25883 29.6 4.2
YKR009c 28.1 9.7
> At5g42890
Length=123
Score = 79.3 bits (194), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 42/120 (35%), Positives = 70/120 (58%), Gaps = 3/120 (2%)
Query 71 AGSCSKAEKIFKMLAAYLELDKEKTQQLKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQDG 130
A + K++ I M+ +L D K ++ K+ + IA K+ + + +DL++ G
Sbjct 2 ANTQLKSDAIMDMMKEHLSTDAGK--EVTEKIGLVYQINIAPKKLGFEEVTYIVDLKK-G 58
Query 131 EGCAKAGVWGAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAAAKFTPQLFP 190
E G DATF+ ++ F ++ G++NPQ+AF++G MKIKGS++AA KFTP +FP
Sbjct 59 EVTKGKYEGGKVDATFSFKDDDFVKVATGKMNPQMAFIRGAMKIKGSLSAAQKFTPDIFP 118
> Hs20559667
Length=156
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 35/94 (37%), Positives = 51/94 (54%), Gaps = 4/94 (4%)
Query 92 KEKTQQLKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQDGEGCAKAG-VWGAADATFAVGE 150
+E QL KVN+ F +I + + +L+WTIDL+ +G G G AD F + E
Sbjct 56 REVGAQLVKKVNAVFQLDIT--KNGKTILRWTIDLK-NGSGDMYPGPARLPADTVFTIPE 112
Query 151 EAFCRICLGELNPQVAFLQGKMKIKGSMAAAAKF 184
F + LG++NPQ AFL GK K+ G + + K
Sbjct 113 SVFMELVLGKMNPQKAFLAGKFKVSGKVLLSWKL 146
> 7292920
Length=115
Score = 55.5 bits (132), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 37/102 (36%), Positives = 55/102 (53%), Gaps = 8/102 (7%)
Query 90 LDKEKTQQLKTK-VNSCFLFEIAAKQQQQPVLQWTIDLRQDG--EGCAKAGVWGAADATF 146
+D K + K K VN FL++I + + +WT+D + EG A+ G+ D T
Sbjct 13 IDGLKENEAKAKAVNGVFLYKIT--KDGKVAKEWTLDCKNAKAYEGPAQ-GI--KVDTTL 67
Query 147 AVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAAAKFTPQL 188
V +E I LG+LNPQ AF++GK+KI G++ K P L
Sbjct 68 TVADEDMVDIALGKLNPQAAFMKGKLKIAGNIMLTQKLAPLL 109
> Hs4504505
Length=736
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 38/158 (24%), Positives = 72/158 (45%), Gaps = 13/158 (8%)
Query 27 DKVSYPSSLQDSLIMASQQIVSEGKAAASGEEGAAAPAPAAAAAAGSCSKAEKIFKMLAA 86
+++ + + +Q++ ++S + G +A P+ S E+I + L
Sbjct 582 NRIHFQTKVQET----GDIVISNAYVDLAPTSGTSAKTPSEGGKLQSTFVFEEIGRRL-- 635
Query 87 YLELDKEKTQQLKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQDGEGCAKAGVWGAADATF 146
K+ ++ KVN+ F + I +WTIDL+ + GAAD T
Sbjct 636 -----KDIGPEVVKKVNAVFEWHITKGGNIGA--KWTIDLKSGSGKVYQGPAKGAADTTI 688
Query 147 AVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAAAKF 184
+ +E F + LG+L+PQ AF G++K +G++ + K
Sbjct 689 ILSDEDFMEVVLGKLDPQKAFFSGRLKARGNIMLSQKL 726
> CE01725
Length=118
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 57/119 (47%), Gaps = 15/119 (12%)
Query 76 KAEKIFKMLAAYLELDKEKTQQLKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQD----GE 131
K++ IF+ + + DKE + KV + F IA + V WTID + D G+
Sbjct 4 KSDVIFEEIKERIATDKEMVK----KVGTSFRMTIAGADGKTKV--WTIDAKSDTPYVGD 57
Query 132 GCAKAGVWGAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAAAKFTPQLFP 190
++ + + + F I G++ P AF+QGKMK+KG++A A K L P
Sbjct 58 DSSRP-----VEIEINIKDSDFIAIAAGKMKPDQAFMQGKMKLKGNIAKAMKLRTILDP 111
> Hs14150062_2
Length=133
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 56/122 (45%), Gaps = 17/122 (13%)
Query 63 PAPAAAAAAGSCSKAEKIFKMLAAYLELDKEKTQQLKTKVNSCFLFEIAAKQQQQPVLQW 122
P P + A E+ F+++ L D K Q + +LFE++ + W
Sbjct 18 PKPRSGAV-------EETFRIVKDSLSDDVVKATQ------AIYLFELSGEDGGT----W 60
Query 123 TIDLRQDGEGCAKAGVWGAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAAA 182
+DL+ G AD ++ + F ++ G+L P +AF+ GK+KIKG+MA A
Sbjct 61 FLDLKSKGGNVGYGEPSDQADVVMSMTTDDFVKMFSGKLKPTMAFMSGKLKIKGNMALAI 120
Query 183 KF 184
K
Sbjct 121 KL 122
> Hs19923233_2
Length=115
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 35/63 (55%), Gaps = 1/63 (1%)
Query 122 WTIDLRQDGEGCAKAGVWGAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAA 181
W +D++ +G+G AD T + + F + G++NPQ AF QGK+KI G+M A
Sbjct 42 WVVDVK-NGKGSVLPNSDKKADCTITMADSDFLALMTGKMNPQSAFFQGKLKITGNMGLA 100
Query 182 AKF 184
K
Sbjct 101 MKL 103
> 7298493_2
Length=123
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 34/109 (31%), Positives = 53/109 (48%), Gaps = 5/109 (4%)
Query 76 KAEKIFKMLAAYLELDKEKTQQLKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQDGEGCAK 135
K + K+L ++ DK+ L KV + + F++ Q W ID +Q G+G
Sbjct 2 KVAPLLKLLEQAMQEDKD---NLIEKVRAIYGFKVNNGPNGQTGF-WVIDAKQ-GKGKII 56
Query 136 AGVWGAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAAAKF 184
D TF + ++ + G+L PQ AF QGK+KI+G+M A K
Sbjct 57 FNGTQKCDVTFIISDDDVFELLTGKLPPQKAFFQGKIKIQGNMGFAMKL 105
> 7301617_2
Length=110
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 39/88 (44%), Gaps = 4/88 (4%)
Query 97 QLKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQDGEGCAKAGVWGAADATFAVGEEAFCRI 156
++ +K + F F I+ +Q W +DL+ C A DAT + + F +
Sbjct 16 EIVSKTQAVFQFNISGAEQGT----WFLDLKNGSGSCGAGTPTAAPDATLTMNSKNFFDM 71
Query 157 CLGELNPQVAFLQGKMKIKGSMAAAAKF 184
G+L A++ GK+KI G A K
Sbjct 72 FSGKLKAAPAYMTGKLKISGDFQKALKL 99
> CE04770
Length=436
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 17/37 (45%), Positives = 25/37 (67%), Gaps = 0/37 (0%)
Query 142 ADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSM 178
A+AT V + F I G+LN Q AF+ GK+K+KG++
Sbjct 382 ANATVTVADSDFVDIAAGKLNAQKAFMSGKLKVKGNV 418
> CE17123_2
Length=91
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 30/61 (49%), Gaps = 1/61 (1%)
Query 124 IDLRQDGEGCAKAGVWGAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAAAK 183
IDL+ G G A G D F E F +I E++P ++ G +K+KGS+ A +
Sbjct 21 IDLKH-GSGSAYKGTSLNPDVVFETSLEVFGKILTKEVSPVTVYMNGNLKVKGSIQDAMQ 79
Query 184 F 184
Sbjct 80 L 80
> CE02490_2
Length=116
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 31/63 (49%), Gaps = 2/63 (3%)
Query 123 TIDLRQDGEG-CAKAGVWGAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAA 181
T+DL+ +GEG G AD F + E F + G+L P A + K++I G M A
Sbjct 43 TLDLK-NGEGALTDKKASGKADVKFTLAPEHFAPLFTGKLRPTTALMTKKLQISGDMPGA 101
Query 182 AKF 184
K
Sbjct 102 MKL 104
> Hs20149563_2
Length=93
Score = 36.6 bits (83), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 40/88 (45%), Gaps = 4/88 (4%)
Query 98 LKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQDGEGCAKAGVW-GAADATFAVGEEAFCRI 156
L ++V +C+ F + Q + +DL G G GV G D + E +
Sbjct 1 LVSQVGACYQFNVVLPSGTQS--AYFLDL-TTGRGRVGHGVPDGIPDVVVEMAEADLRAL 57
Query 157 CLGELNPQVAFLQGKMKIKGSMAAAAKF 184
EL P A++ G++K+KG +A A K
Sbjct 58 LCRELRPLGAYMSGRLKVKGDLAMAMKL 85
> Hs22050448
Length=421
Score = 35.8 bits (81), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 28/57 (49%), Gaps = 0/57 (0%)
Query 26 KDKVSYPSSLQDSLIMASQQIVSEGKAAASGEEGAAAPAPAAAAAAGSCSKAEKIFK 82
+D+ ++P L+ ++ Q SEGK E GAA P ++ AEK+FK
Sbjct 104 EDERAHPEHLKSYRVIQHQDTHSEGKPRRHTEHGAAFPPGSSCGQQQEVHVAEKLFK 160
> YMR286w
Length=86
Score = 29.6 bits (65), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 37/81 (45%), Gaps = 1/81 (1%)
Query 29 VSYPSSLQDSLIMASQQIVSEGKAAASGEEGAAAPAPAAAAAAGSCSKAEKIFKMLAAYL 88
V Y +L SLI S K+ G+ G+ A AGS +K +++ K+
Sbjct 2 VFYKVTLSRSLIGVPHTTKSIVKSLGLGKRGSIVYKKVNPAIAGSLAKVKELVKVEVTEH 61
Query 89 ELDKEKTQQLKTKVNSCFLFE 109
EL + ++L+ K N F+ E
Sbjct 62 ELTPSQQRELR-KSNPGFIVE 81
> Hs14752790
Length=1034
Score = 29.6 bits (65), Expect = 3.8, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 17/34 (50%), Gaps = 0/34 (0%)
Query 47 VSEGKAAASGEEGAAAPAPAAAAAAGSCSKAEKI 80
V+ GKA S E P P +A AG CS KI
Sbjct 778 VNAGKAGPSAEAPKVHPKPNSAPVAGPCSSFPKI 811
> CE25883
Length=1488
Score = 29.6 bits (65), Expect = 4.2, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 27/46 (58%), Gaps = 4/46 (8%)
Query 99 KTKVNSCFLFEIAAKQQQQPVLQWTI-DLRQDGEGCAK--AGVWGA 141
KT +N CFL ++AA ++Q+ +L W + + G C K G WG+
Sbjct 336 KTYLNECFL-KLAACKEQKDILVWKRGNCDEAGSPCEKMECGFWGS 380
> YKR009c
Length=900
Score = 28.1 bits (61), Expect = 9.7, Method: Composition-based stats.
Identities = 14/58 (24%), Positives = 28/58 (48%), Gaps = 9/58 (15%)
Query 7 PSSFTLEDLSKEWKTVLDFKD----KVSYPSSLQDSLIMASQQIVSEGKAAASGEEGA 60
P ++T E + +WK + D++D K +P L D ++++ K E+G+
Sbjct 262 PKTYTPEAILNKWKEITDYRDKPFNKTQHPYQLSD-----YNDLITKAKKLPPNEQGS 314
Lambda K H
0.315 0.128 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3166472848
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40