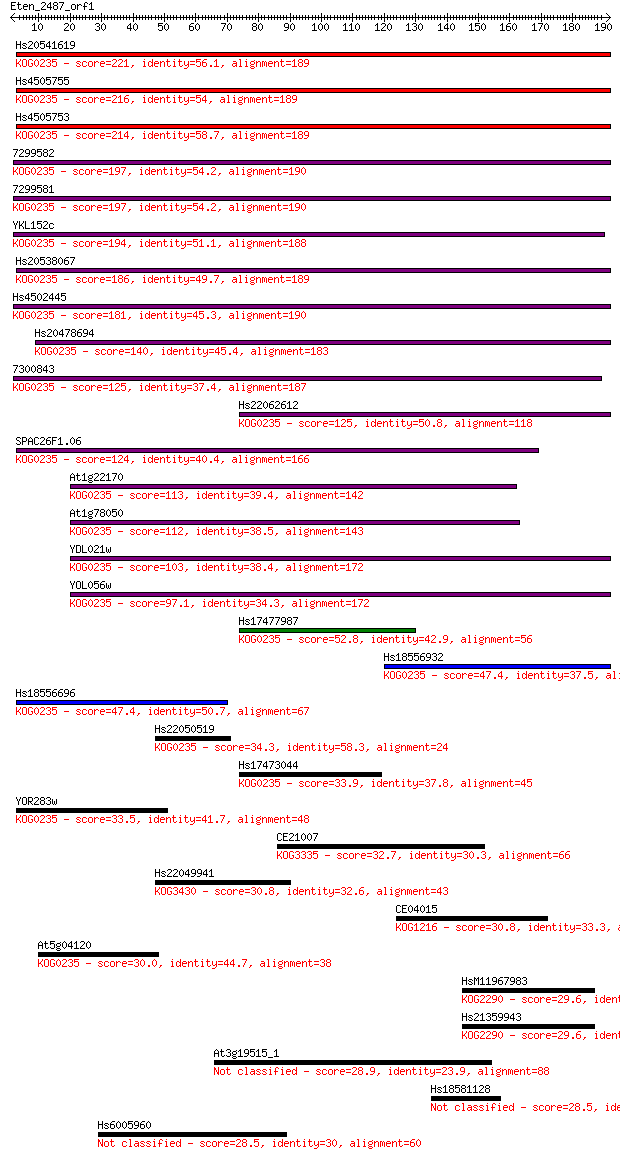
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2487_orf1
Length=191
Score E
Sequences producing significant alignments: (Bits) Value
Hs20541619 221 6e-58
Hs4505755 216 3e-56
Hs4505753 214 1e-55
7299582 197 8e-51
7299581 197 9e-51
YKL152c 194 1e-49
Hs20538067 186 2e-47
Hs4502445 181 8e-46
Hs20478694 140 2e-33
7300843 125 6e-29
Hs22062612 125 7e-29
SPAC26F1.06 124 1e-28
At1g22170 113 3e-25
At1g78050 112 7e-25
YDL021w 103 2e-22
YOL056w 97.1 2e-20
Hs17477987 52.8 5e-07
Hs18556932 47.4 2e-05
Hs18556696 47.4 2e-05
Hs22050519 34.3 0.14
Hs17473044 33.9 0.20
YOR283w 33.5 0.28
CE21007 32.7 0.44
Hs22049941 30.8 1.7
CE04015 30.8 1.8
At5g04120 30.0 3.2
HsM11967983 29.6 3.7
Hs21359943 29.6 3.7
At3g19515_1 28.9 6.3
Hs18581128 28.5 7.8
Hs6005960 28.5 8.3
> Hs20541619
Length=253
Score = 221 bits (564), Expect = 6e-58, Method: Compositional matrix adjust.
Identities = 106/192 (55%), Positives = 140/192 (72%), Gaps = 3/192 (1%)
Query 3 RAVETCWTVLMHSNQCYLPIKNTWRLNERHYGALQGLNKAETAAKHGEEQVKIWRRAYAV 62
RA+ T W +L ++Q +LP+ TWRLNERHYG L GLNKAETAAKHGEEQVKIWRR++ +
Sbjct 62 RAIRTLWAILDGTDQMWLPVVRTWRLNERHYGGLTGLNKAETAAKHGEEQVKIWRRSFDI 121
Query 63 PPPPLDPEDK--RNAKFEEKYKHLPQEVLPLTECLKDTVERVLPYYFDEIAPALLDNKKV 120
PPPP+D + + E +Y L LP E LKDT+ R LP++ +EI P + K+V
Sbjct 122 PPPPMDEKHPYYNSISKERRYAGLKPGELPTCESLKDTIARALPFWNEEIVPQIKAGKRV 181
Query 121 LVVAHGNSLRGLVKHLDGMSEEEVLELNIPTAVPLVYELDESLKPLKKY-YLLDEAEVQK 179
L+ AHGNSLRG+VKHL+GMS++ ++ELN+PT +P+VYEL++ LKP K +L DE V+K
Sbjct 182 LIAAHGNSLRGIVKHLEGMSDQAIMELNLPTGIPIVYELNKELKPTKPMQFLGDEETVRK 241
Query 180 RIAAVANQGKAK 191
+ AVA QGKAK
Sbjct 242 AMEAVAAQGKAK 253
> Hs4505755
Length=253
Score = 216 bits (549), Expect = 3e-56, Method: Compositional matrix adjust.
Identities = 102/192 (53%), Positives = 137/192 (71%), Gaps = 3/192 (1%)
Query 3 RAVETCWTVLMHSNQCYLPIKNTWRLNERHYGALQGLNKAETAAKHGEEQVKIWRRAYAV 62
RA+ T W +L ++Q +LP+ TWR NERHYG L G NKAETAAKHGEEQV+ WRR++ +
Sbjct 62 RAIRTLWAILDGTDQMWLPVVRTWRFNERHYGGLTGFNKAETAAKHGEEQVRSWRRSFDI 121
Query 63 PPPPLDPEDK--RNAKFEEKYKHLPQEVLPLTECLKDTVERVLPYYFDEIAPALLDNKKV 120
PPPP+D + + E +Y L LP E LKDT+ R LP++ +EI P + K+V
Sbjct 122 PPPPMDEKHPYYNSISKERRYAGLKPGELPTCESLKDTIARALPFWNEEIVPQIKAGKRV 181
Query 121 LVVAHGNSLRGLVKHLDGMSEEEVLELNIPTAVPLVYELDESLKPLKKY-YLLDEAEVQK 179
L+ AHGNSLRG+VKHL+GMS++ ++ELN+PT +P+VYEL++ LKP K +L DE V+K
Sbjct 182 LIAAHGNSLRGIVKHLEGMSDQAIMELNLPTGIPIVYELNKELKPTKPMQFLGDEETVRK 241
Query 180 RIAAVANQGKAK 191
+ AVA QGKAK
Sbjct 242 AMEAVAAQGKAK 253
> Hs4505753
Length=254
Score = 214 bits (544), Expect = 1e-55, Method: Compositional matrix adjust.
Identities = 111/192 (57%), Positives = 145/192 (75%), Gaps = 3/192 (1%)
Query 3 RAVETCWTVLMHSNQCYLPIKNTWRLNERHYGALQGLNKAETAAKHGEEQVKIWRRAYAV 62
RA+ T WTVL +Q +LP+ TWRLNERHYG L GLNKAETAAKHGE QVKIWRR+Y V
Sbjct 62 RAIRTLWTVLDAIDQMWLPVVRTWRLNERHYGGLTGLNKAETAAKHGEAQVKIWRRSYDV 121
Query 63 PPPPLDPEDK--RNAKFEEKYKHLPQEVLPLTECLKDTVERVLPYYFDEIAPALLDNKKV 120
PPPP++P+ N + +Y L ++ LP E LKDT+ R LP++ +EI P + + K+V
Sbjct 122 PPPPMEPDHPFYSNISKDRRYADLTEDQLPSCESLKDTIARALPFWNEEIVPQIKEGKRV 181
Query 121 LVVAHGNSLRGLVKHLDGMSEEEVLELNIPTAVPLVYELDESLKPLK-KYYLLDEAEVQK 179
L+ AHGNSLRG+VKHL+G+SEE ++ELN+PT +P+VYELD++LKP+K +L DE V+K
Sbjct 182 LIAAHGNSLRGIVKHLEGLSEEAIMELNLPTGIPIVYELDKNLKPIKPMQFLGDEETVRK 241
Query 180 RIAAVANQGKAK 191
+ AVA QGKAK
Sbjct 242 AMEAVAAQGKAK 253
> 7299582
Length=292
Score = 197 bits (502), Expect = 8e-51, Method: Compositional matrix adjust.
Identities = 103/194 (53%), Positives = 134/194 (69%), Gaps = 4/194 (2%)
Query 2 TRAVETCWTVLMHSNQCYLPIKNTWRLNERHYGALQGLNKAETAAKHGEEQVKIWRRAYA 61
TRA ET L S +P+ TWRLNERHYG L GLNKAETA K GEE+VKIWRR++
Sbjct 99 TRAQETLRAALKSSEHKKIPVCTTWRLNERHYGGLTGLNKAETAKKFGEEKVKIWRRSFD 158
Query 62 VPPPPLDPEDKRNAKFEE--KYK-HLPQEVLPLTECLKDTVERVLPYYFDEIAPALLDNK 118
PPPP++ + + A E +YK L E P +E LK T+ER LPY+ + I P + D
Sbjct 159 TPPPPMEKDHEYYACIVEDPRYKDQLKPEEFPKSESLKLTIERTLPYWNEVIVPQIKDGM 218
Query 119 KVLVVAHGNSLRGLVKHLDGMSEEEVLELNIPTAVPLVYELDESLKPLKKYYLLDEAE-V 177
+VL+ AHGNSLRG+VKHL+ +S+++++ LN+PT +P VYELDESLKPL L + E V
Sbjct 219 RVLIAAHGNSLRGVVKHLECISDKDIMSLNLPTGIPFVYELDESLKPLATLKFLGDPETV 278
Query 178 QKRIAAVANQGKAK 191
+K + +VANQGKAK
Sbjct 279 KKAMESVANQGKAK 292
> 7299581
Length=271
Score = 197 bits (501), Expect = 9e-51, Method: Compositional matrix adjust.
Identities = 103/194 (53%), Positives = 134/194 (69%), Gaps = 4/194 (2%)
Query 2 TRAVETCWTVLMHSNQCYLPIKNTWRLNERHYGALQGLNKAETAAKHGEEQVKIWRRAYA 61
TRA ET L S +P+ TWRLNERHYG L GLNKAETA K GEE+VKIWRR++
Sbjct 78 TRAQETLRAALKSSEHKKIPVCTTWRLNERHYGGLTGLNKAETAKKFGEEKVKIWRRSFD 137
Query 62 VPPPPLDPEDKRNAKFEE--KYK-HLPQEVLPLTECLKDTVERVLPYYFDEIAPALLDNK 118
PPPP++ + + A E +YK L E P +E LK T+ER LPY+ + I P + D
Sbjct 138 TPPPPMEKDHEYYACIVEDPRYKDQLKPEEFPKSESLKLTIERTLPYWNEVIVPQIKDGM 197
Query 119 KVLVVAHGNSLRGLVKHLDGMSEEEVLELNIPTAVPLVYELDESLKPLKKYYLLDEAE-V 177
+VL+ AHGNSLRG+VKHL+ +S+++++ LN+PT +P VYELDESLKPL L + E V
Sbjct 198 RVLIAAHGNSLRGVVKHLECISDKDIMSLNLPTGIPFVYELDESLKPLATLKFLGDPETV 257
Query 178 QKRIAAVANQGKAK 191
+K + +VANQGKAK
Sbjct 258 KKAMESVANQGKAK 271
> YKL152c
Length=247
Score = 194 bits (492), Expect = 1e-49, Method: Compositional matrix adjust.
Identities = 96/188 (51%), Positives = 132/188 (70%), Gaps = 0/188 (0%)
Query 2 TRAVETCWTVLMHSNQCYLPIKNTWRLNERHYGALQGLNKAETAAKHGEEQVKIWRRAYA 61
+RA++T L +++ ++P+ +WRLNERHYG LQG +KAET K GEE+ +RR++
Sbjct 59 SRAIQTANIALEKADRLWIPVNRSWRLNERHYGDLQGKDKAETLKKFGEEKFNTYRRSFD 118
Query 62 VPPPPLDPEDKRNAKFEEKYKHLPQEVLPLTECLKDTVERVLPYYFDEIAPALLDNKKVL 121
VPPPP+D + K +E+YK++ VLP TE L ++R+LPY+ D IA LL K V+
Sbjct 119 VPPPPIDASSPFSQKGDERYKYVDPNVLPETESLALVIDRLLPYWQDVIAKDLLSGKTVM 178
Query 122 VVAHGNSLRGLVKHLDGMSEEEVLELNIPTAVPLVYELDESLKPLKKYYLLDEAEVQKRI 181
+ AHGNSLRGLVKHL+G+S+ ++ +LNIPT +PLV+ELDE+LKP K Y LD
Sbjct 179 IAAHGNSLRGLVKHLEGISDADIAKLNIPTGIPLVFELDENLKPSKPSYYLDPEAAAAGA 238
Query 182 AAVANQGK 189
AAVANQGK
Sbjct 239 AAVANQGK 246
> Hs20538067
Length=224
Score = 186 bits (473), Expect = 2e-47, Method: Compositional matrix adjust.
Identities = 94/190 (49%), Positives = 134/190 (70%), Gaps = 12/190 (6%)
Query 3 RAVETCWTVLMHSNQCYLPIKNTWRLNERHYGALQGLNKAETAAKHGEEQVKIWRRAYAV 62
R ++T WTVL +Q +LP+ H G L GLNKAETAAKHGE QVKIWR +Y V
Sbjct 45 RVIQTLWTVLDAIDQTWLPV---------HSGGLIGLNKAETAAKHGEAQVKIWRHSYDV 95
Query 63 PPPPLDPEDK-RNAKFEEKYKHLPQEVLPLTECLKDTVERVLPYYFDEIAPALLDNKKVL 121
PPP ++P+ N + ++ +L ++ LP E LKDT+ + LP++ +EI P + + K+VL
Sbjct 96 PPPLMEPDHPFYNISKDRRFANLTEDQLPSCESLKDTIAKALPFWNEEIVPQIKEGKQVL 155
Query 122 VVAHGNSLRGLVKHLDGMSEEEVLELNIPTAVPLVYELDESLKPLKKYYLLDEAEVQKRI 181
+ AHGNS G+VKHL+G+SEE ++ELN+PT +P+VYELD++LKP++ +L DE ++K +
Sbjct 156 IAAHGNSPPGIVKHLEGLSEEAIMELNLPTGIPVVYELDKNLKPIQ--FLGDEETMRKAM 213
Query 182 AAVANQGKAK 191
AVA QGKAK
Sbjct 214 EAVAAQGKAK 223
> Hs4502445
Length=259
Score = 181 bits (459), Expect = 8e-46, Method: Compositional matrix adjust.
Identities = 86/195 (44%), Positives = 134/195 (68%), Gaps = 5/195 (2%)
Query 2 TRAVETCWTVLMHSNQCYLPIKNTWRLNERHYGALQGLNKAETAAKHGEEQVKIWRRAYA 61
R++ T W +L Q ++P++++WRLNERHYGAL GLN+ + A HGEEQV++WRR+Y
Sbjct 61 NRSIHTAWLILEELGQEWVPVESSWRLNERHYGALIGLNREQMALNHGEEQVRLWRRSYN 120
Query 62 VPPPPLDPEDKRNAKF--EEKYK--HLPQEVLPLTECLKDTVERVLPYYFDEIAPALLDN 117
V PPP++ + + +YK +P + LP +E LKD +ER+LPY+ + IAP +L
Sbjct 121 VTPPPIEESHPYYQEIYNDRRYKVCDVPLDQLPRSESLKDVLERLLPYWNERIAPEVLRG 180
Query 118 KKVLVVAHGNSLRGLVKHLDGMSEEEVLELNIPTAVPLVYELDESLKPLKKY-YLLDEAE 176
K +L+ AHGNS R L+KHL+G+S+E+++ + +PT VP++ ELDE+L+ + + +L D+
Sbjct 181 KTILISAHGNSSRALLKHLEGISDEDIINITLPTGVPILLELDENLRAVGPHQFLGDQEA 240
Query 177 VQKRIAAVANQGKAK 191
+Q I V +QGK K
Sbjct 241 IQAAIKKVEDQGKVK 255
> Hs20478694
Length=250
Score = 140 bits (352), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 83/184 (45%), Positives = 107/184 (58%), Gaps = 33/184 (17%)
Query 9 WTVLMHSNQCYLPIKNTWRLNERHYGALQGLNKAETAAKHGEEQVKIWRRAYAVPPPPLD 68
WTVL +Q LP+ TW L E HYG+L GLNKAET+AKHGE Q I +
Sbjct 60 WTVLDAIDQMRLPVVRTWCLKEWHYGSLAGLNKAETSAKHGEAQSNISKGC--------- 110
Query 69 PEDKRNAKFEEKYKHLPQEVLPLTECLKDTVERVLPYYFDEIAPALLDNKKVLVVAHGNS 128
+Y L ++ L E LK EI P + + K VL AHGNS
Sbjct 111 -----------RYADLTEDQLSSGESLK------------EIVPQIKEGKWVLTAAHGNS 147
Query 129 LRGLVKHLDGMSEEEVLELNIPTAVPLVYELDESLKPLKKY-YLLDEAEVQKRIAAVANQ 187
LRG+VKHL+G+SEE ++ELN+PT +P+VYELD++LKP+K +L DE + K + AVA Q
Sbjct 148 LRGIVKHLEGLSEEAIMELNLPTGIPIVYELDKNLKPIKPMQFLGDEVTLCKAMEAVAAQ 207
Query 188 GKAK 191
KAK
Sbjct 208 SKAK 211
> 7300843
Length=253
Score = 125 bits (313), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 70/191 (36%), Positives = 103/191 (53%), Gaps = 5/191 (2%)
Query 2 TRAVETCWTVLMHSNQCYLPIKNTWRLNERHYGALQGLNKAETAAKHGEEQVKIWRRAYA 61
+R+ +T +L N Y+PIK WRL ERHYG L G K A ++GEEQV+ WRR Y
Sbjct 63 SRSRQTAELILSKLNCAYVPIKEDWRLCERHYGNLTGCRKRVVADRYGEEQVQAWRRGYD 122
Query 62 VPPPPLDPEDKRNAKFEEK--YKHLPQEVLPLTECLKDTVERVLPYYFDEIAPALLDNKK 119
PPP+D +++ + +P+ PL E L V+RV P + E+ + +
Sbjct 123 CVPPPIDEKNRYFYTICSNPIFDDVPRGEFPLAESLHMCVDRVKPVW-KEVRREVFQGTR 181
Query 120 VLVVAHGNSLRGLVKHLDGMSEEEVLELNIPTAVPLVYELDESLKPL--KKYYLLDEAEV 177
VL+ HG R LV+H++G+S E + ++NIP VP VYE D L L D+ +
Sbjct 182 VLMCVHGTVARALVQHIEGISNEAIEKVNIPNCVPRVYEFDLKTGGLVGAAINLGDQEYI 241
Query 178 QKRIAAVANQG 188
+++ A VA G
Sbjct 242 RRKTAQVAAIG 252
> Hs22062612
Length=129
Score = 125 bits (313), Expect = 7e-29, Method: Compositional matrix adjust.
Identities = 60/119 (50%), Positives = 87/119 (73%), Gaps = 1/119 (0%)
Query 74 NAKFEEKYKHLPQEVLPLTECLKDTVERVLPYYFDEIAPALLDNKKVLVVAHGNSLRGLV 133
N + +Y L +++LP E LKDT+ R L ++ +EI P + + K+VL+ AHGNSLRG+
Sbjct 10 NISKDRRYADLTEDLLPSCESLKDTIARALTFWNEEIVPQIKEGKRVLIAAHGNSLRGIA 69
Query 134 KHLDGMSEEEVLELNIPTAVPLVYELDESLKPLKKY-YLLDEAEVQKRIAAVANQGKAK 191
KHL+G+SEE ++ELN+PT +P+VYE D++LKP+K +L DE V+K + AVA QGKAK
Sbjct 70 KHLEGLSEEAIMELNLPTGIPIVYEWDKNLKPIKPMQFLGDEEMVRKAMEAVAAQGKAK 128
> SPAC26F1.06
Length=211
Score = 124 bits (312), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 67/166 (40%), Positives = 93/166 (56%), Gaps = 25/166 (15%)
Query 3 RAVETCWTVLMHSNQCYLPIKNTWRLNERHYGALQGLNKAETAAKHGEEQVKIWRRAYAV 62
RA +TC +L + L + +LNER+YG LQGLNK + K G EQV+IWRR+Y +
Sbjct 66 RAQKTCQIILEEVGEPNLETIKSEKLNERYYGDLQGLNKDDARKKWGAEQVQIWRRSYDI 125
Query 63 PPPPLDPEDKRNAKFEEKYKHLPQEVLPLTECLKDTVERVLPYYFDEIAPALLDNKKVLV 122
PP E LKDT ERVLPYY I P +L +KVL+
Sbjct 126 APPN-------------------------GESLKDTAERVLPYYKSTIVPHILKGEKVLI 160
Query 123 VAHGNSLRGLVKHLDGMSEEEVLELNIPTAVPLVYELDESLKPLKK 168
AHGNSLR L+ L+G++ +++++ + T VP+VY LD+ K + K
Sbjct 161 AAHGNSLRALIMDLEGLTGDQIVKRELATGVPIVYHLDKDGKYVSK 206
> At1g22170
Length=334
Score = 113 bits (282), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 56/142 (39%), Positives = 79/142 (55%), Gaps = 25/142 (17%)
Query 20 LPIKNTWRLNERHYGALQGLNKAETAAKHGEEQVKIWRRAYAVPPPPLDPEDKRNAKFEE 79
+P+ W+LNER YG LQGLNK ETA ++G+EQV WRR+Y +PPP
Sbjct 178 IPVIPAWQLNERMYGELQGLNKQETAERYGKEQVHEWRRSYDIPPPK------------- 224
Query 80 KYKHLPQEVLPLTECLKDTVERVLPYYFDEIAPALLDNKKVLVVAHGNSLRGLVKHLDGM 139
E L+ ER + Y+ D I P L K V++ AHGNSLR ++ +LD +
Sbjct 225 ------------GESLEMCAERAVAYFQDNIEPKLAAGKNVMIAAHGNSLRSIIMYLDKL 272
Query 140 SEEEVLELNIPTAVPLVYELDE 161
+ +EV+ L + T +PL+Y E
Sbjct 273 TCQEVISLELSTGIPLLYIFKE 294
> At1g78050
Length=195
Score = 112 bits (279), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 55/143 (38%), Positives = 77/143 (53%), Gaps = 25/143 (17%)
Query 20 LPIKNTWRLNERHYGALQGLNKAETAAKHGEEQVKIWRRAYAVPPPPLDPEDKRNAKFEE 79
+P+ W+LNER YG LQGLNK ETA ++G +QV WRR+Y +PPP
Sbjct 42 IPVIAAWQLNERMYGELQGLNKKETAERYGTQQVHEWRRSYEIPPPK------------- 88
Query 80 KYKHLPQEVLPLTECLKDTVERVLPYYFDEIAPALLDNKKVLVVAHGNSLRGLVKHLDGM 139
E L+ ER + Y+ D I P L V++ AHGNSLR ++ +LD +
Sbjct 89 ------------GESLEMCAERAVAYFEDNIKPELASGNNVMIAAHGNSLRSIIMYLDDL 136
Query 140 SEEEVLELNIPTAVPLVYELDES 162
+ +EV L++ T VPL+Y E
Sbjct 137 TSQEVTTLDLSTGVPLLYIFKEG 159
> YDL021w
Length=311
Score = 103 bits (257), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 66/195 (33%), Positives = 108/195 (55%), Gaps = 24/195 (12%)
Query 20 LPIKNTWRLNERHYGALQGLNKAETAAKHGEEQVKIWRRAYAVPPPPLD-------PEDK 72
+PI TWRLNERHYG+ QG K ++G+++ RR Y PPP+D E++
Sbjct 116 IPILQTWRLNERHYGSWQGQRKPNVLKEYGKDKYMFIRRDYEGKPPPVDLDREMIQQENE 175
Query 73 RNA----KFEEKYKHLPQE--------VLPLTECLKDTVERVLPYYFDEI--APALLDNK 118
+ + +F+E + + E VLP +E L++ V R+ P+ + I D
Sbjct 176 KGSSTGYEFKEPNRQIKYELECSNHDIVLPDSESLREVVYRLNPFLQNVILKLANQYDES 235
Query 119 KVLVVAHGNSLRGLVKHLDGMSEEEVLELNIPTAVPLVYELDES--LKPLKKYYLLDEAE 176
L+V HG+S+R L+K L+G+S++++ ++IP +PLV ELD++ LK ++K+Y LD
Sbjct 236 SCLIVGHGSSVRSLLKILEGISDDDIKNVDIPNGIPLVVELDKNNGLKFIRKFY-LDPES 294
Query 177 VQKRIAAVANQGKAK 191
+ V N+G K
Sbjct 295 AKINAEKVRNEGFIK 309
> YOL056w
Length=303
Score = 97.1 bits (240), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 59/192 (30%), Positives = 96/192 (50%), Gaps = 20/192 (10%)
Query 20 LPIKNTWRLNERHYGALQGLNKAETAAKHGEEQVKIWRRAYAVPPPPL-----------D 68
+P+ TWRLNERHYGA QG K + ++G+E+ RR Y PP + D
Sbjct 110 MPVLQTWRLNERHYGAWQGQRKPDILKEYGKEKYMYIRRDYNGKPPKVNLNLEMVQEEND 169
Query 69 PEDKRNAKFEEKYKHL-------PQEVLPLTECLKDTVERVLPYYFDEIAPAL--LDNKK 119
F+E +HL E LP +E L + V R+ P+ + + + +
Sbjct 170 QGSSTGYDFKEPNRHLKYGPEEKANERLPESESLCEVVVRLKPFLNNVVLSTANKISQES 229
Query 120 VLVVAHGNSLRGLVKHLDGMSEEEVLELNIPTAVPLVYELDESLKPLKKYYLLDEAEVQK 179
++V HG+S+R L+K L+G+S+E++ +++IP +PLV ELD + + LD +
Sbjct 230 CVIVGHGSSVRSLLKVLEGISDEDIKDVDIPNGIPLVIELDRDNYSFVRKFYLDPESAKV 289
Query 180 RIAAVANQGKAK 191
V ++G K
Sbjct 290 NAQMVRDEGFEK 301
> Hs17477987
Length=152
Score = 52.8 bits (125), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 24/56 (42%), Positives = 35/56 (62%), Gaps = 0/56 (0%)
Query 74 NAKFEEKYKHLPQEVLPLTECLKDTVERVLPYYFDEIAPALLDNKKVLVVAHGNSL 129
N + +Y L ++ LP LKDT+ R LP++ +EI P + + K VL+ AHGNSL
Sbjct 97 NVSKDRRYADLTEDQLPSCNSLKDTLARALPFWNEEIVPQIKEGKWVLIAAHGNSL 152
> Hs18556932
Length=97
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 48/74 (64%), Gaps = 4/74 (5%)
Query 120 VLVVAHGNSLRGLVKHLDGMSEEEVLELNIPTAVPLVYELD--ESLKPLKKYYLLDEAEV 177
+++ NSL+G+VKH +G SEE +EL++ T++ +VYEL+ +S K ++ YL ++ +
Sbjct 25 LILFTRKNSLQGIVKHREGCSEEAAIELSLQTSILVVYELNWEKSFKHMQ--YLGNKEAM 82
Query 178 QKRIAAVANQGKAK 191
+ I AV +G K
Sbjct 83 YEAIQAVTVKGMVK 96
> Hs18556696
Length=300
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 34/67 (50%), Positives = 44/67 (65%), Gaps = 2/67 (2%)
Query 3 RAVETCWTVLMHSNQCYLPIKNTWRLNERHYGALQGLNKAETAAKHGEEQVKIWRRAYAV 62
+A+ T TVL +Q ++P+ TW NE HYG L GLNKAE AAKH E VKIW+ +Y V
Sbjct 232 KAIWTICTVLDAIDQMWMPVMRTW--NECHYGVLIGLNKAEAAAKHEEALVKIWKCSYDV 289
Query 63 PPPPLDP 69
P ++P
Sbjct 290 SSPLMEP 296
> Hs22050519
Length=487
Score = 34.3 bits (77), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 14/24 (58%), Positives = 18/24 (75%), Gaps = 0/24 (0%)
Query 47 KHGEEQVKIWRRAYAVPPPPLDPE 70
+H E QVKIWR +YAV PP +P+
Sbjct 247 QHAEAQVKIWRCSYAVLPPLKEPD 270
> Hs17473044
Length=132
Score = 33.9 bits (76), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 74 NAKFEEKYKHLPQEVLPLTECLKDTVERVLPYYFDEIAPALLDNK 118
N + +Y L ++ LP E LKDT R L ++ +EIAP + + K
Sbjct 88 NISKDGRYADLTEDQLPSCEHLKDTTTRALLFWNEEIAPQIKEGK 132
> YOR283w
Length=230
Score = 33.5 bits (75), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 26/51 (50%), Gaps = 3/51 (5%)
Query 3 RAVETCWTVLMHSNQCYLPIKNTWRLNERHYGALQGLNKAET---AAKHGE 50
R +T VL HS Q +P T L ER+ G ++G+ E A KHGE
Sbjct 75 RCRQTTALVLKHSKQENVPTSYTSGLRERYMGVIEGMQITEAEKYADKHGE 125
> CE21007
Length=266
Score = 32.7 bits (73), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 35/66 (53%), Gaps = 1/66 (1%)
Query 86 QEVLPLTECLKDTVERVLPYYFDEIAPALLDNKKVLVVAHGNSLRGLVKHLDGMSEEEVL 145
+E P TE LK +R+ D A +LD+++ LVV G+S+ G+ ++ + + L
Sbjct 169 EEKKPATESLKQRFQRISSLPIDAAASLILDDEEYLVVRRGSSV-GIELSVNFAARGDRL 227
Query 146 ELNIPT 151
IP+
Sbjct 228 RYRIPS 233
> Hs22049941
Length=109
Score = 30.8 bits (68), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 21/43 (48%), Gaps = 1/43 (2%)
Query 47 KHGEEQVKIWRRAYAVPPPPLDPEDKRNAKFEEKYKHLPQEVL 89
K+G V+IW +A+ VPP P ++ KY+ L L
Sbjct 48 KNGHSSVQIWLKAWTVPPTS-GPSKNKDCSLNSKYQRLKFSAL 89
> CE04015
Length=567
Score = 30.8 bits (68), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 29/48 (60%), Gaps = 2/48 (4%)
Query 124 AHGNSLRGLVKHLDGMSEEEVLELNIPTAVPLVYELDESLKPLKKYYL 171
A G+SL ++K +D SE EVL +N P+ + V + S++ + + Y+
Sbjct 135 AQGDSLPDIMKAMD--SEAEVLGVNCPSDIIFVIDATSSVRGIFEQYI 180
> At5g04120
Length=238
Score = 30.0 bits (66), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 17/39 (43%), Positives = 21/39 (53%), Gaps = 1/39 (2%)
Query 10 TVLMHSNQCYLP-IKNTWRLNERHYGALQGLNKAETAAK 47
T LM + C+ P + L ERH G+LQGL E A K
Sbjct 86 TALMIAKTCFCPEVIEVPDLKERHVGSLQGLYWKEGAEK 124
> HsM11967983
Length=855
Score = 29.6 bits (65), Expect = 3.7, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 145 LELNIPTAVPLVYELDESLKPLKKYYLLDEAEVQKRIAAVAN 186
L+L+IP+AVPL E L+PL++ L + A +++
Sbjct 20 LKLDIPSAVPLTAEEPSFLQPLRRQAFLRSVSMPAETAHISS 61
> Hs21359943
Length=855
Score = 29.6 bits (65), Expect = 3.7, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 145 LELNIPTAVPLVYELDESLKPLKKYYLLDEAEVQKRIAAVAN 186
L+L+IP+AVPL E L+PL++ L + A +++
Sbjct 20 LKLDIPSAVPLTAEEPSFLQPLRRQAFLRSVSMPAETAHISS 61
> At3g19515_1
Length=169
Score = 28.9 bits (63), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 21/94 (22%), Positives = 44/94 (46%), Gaps = 6/94 (6%)
Query 66 PLDPEDKRNAKFEEKYKHLPQEVLPLTECLKDTVERVLPYYFDEIAPALL------DNKK 119
P P D+ F + YK + ++ L + +K +VE+++ D LL D+K
Sbjct 43 PTQPSDRIGDDFSKLYKESTERLINLEDLIKASVEQLVQEMSDHDKAYLLFDSMSGDDKA 102
Query 120 VLVVAHGNSLRGLVKHLDGMSEEEVLELNIPTAV 153
+L + ++ L ++ ++ EVL +P+ +
Sbjct 103 ILYMKRQDTKTELRTCINILAMTEVLHAEVPSFI 136
> Hs18581128
Length=384
Score = 28.5 bits (62), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 10/22 (45%), Positives = 17/22 (77%), Gaps = 0/22 (0%)
Query 135 HLDGMSEEEVLELNIPTAVPLV 156
HL+ + EE ++ELN+ T +P+V
Sbjct 363 HLESLPEEAIMELNLLTGIPIV 384
> Hs6005960
Length=873
Score = 28.5 bits (62), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 18/64 (28%), Positives = 29/64 (45%), Gaps = 4/64 (6%)
Query 29 NERHYGALQGLNKAETAAKHGEEQVKIW----RRAYAVPPPPLDPEDKRNAKFEEKYKHL 84
N+ Y + + AK+ EEQ+KIW R + V P + E+ R +F +
Sbjct 304 NKFPYPTMSEITVLSAQAKYTEEQIKIWFSAQRLKHGVSWTPEEVEEARRKQFNGTVHTV 363
Query 85 PQEV 88
PQ +
Sbjct 364 PQTI 367
Lambda K H
0.315 0.133 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3202455494
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40