bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2493_orf1
Length=135
Score E
Sequences producing significant alignments: (Bits) Value
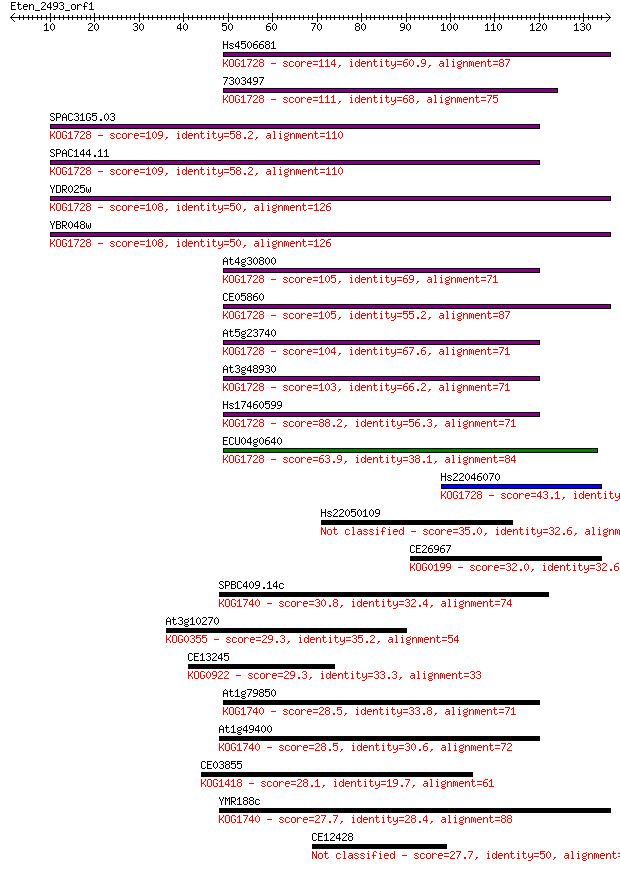
Hs4506681 114 5e-26
7303497 111 3e-25
SPAC31G5.03 109 2e-24
SPAC144.11 109 2e-24
YDR025w 108 4e-24
YBR048w 108 4e-24
At4g30800 105 2e-23
CE05860 105 3e-23
At5g23740 104 4e-23
At3g48930 103 7e-23
Hs17460599 88.2 4e-18
ECU04g0640 63.9 7e-11
Hs22046070 43.1 2e-04
Hs22050109 35.0 0.043
CE26967 32.0 0.37
SPBC409.14c 30.8 0.79
At3g10270 29.3 1.9
CE13245 29.3 2.3
At1g79850 28.5 3.5
At1g49400 28.5 3.5
CE03855 28.1 5.4
YMR188c 27.7 6.5
CE12428 27.7 7.1
> Hs4506681
Length=158
Score = 114 bits (285), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 53/87 (60%), Positives = 66/87 (75%), Gaps = 3/87 (3%)
Query 49 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPL 108
G+V KM+R +VIRR YLH++RKY+R EKRHKN++ HLSPCF V+ GDIVT G+CRPL
Sbjct 75 GVVTKMKMQRTIVIRRDYLHYIRKYNRFEKRHKNMSVHLSPCFRDVQIGDIVTVGECRPL 134
Query 109 SKTIRFNVLKVESNQVFGNVRKQFRLF 135
SKT+RFNVLKV +KQF+ F
Sbjct 135 SKTVRFNVLKVTKA---AGTKKQFQKF 158
> 7303497
Length=155
Score = 111 bits (278), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 51/75 (68%), Positives = 60/75 (80%), Gaps = 0/75 (0%)
Query 49 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPL 108
G+V AKM+R +VIRR YLHFVRKYSR EKRH+N++ H SP F V+ GDIVT G+CRPL
Sbjct 72 GVVRKAKMQRTIVIRRDYLHFVRKYSRFEKRHRNMSVHCSPVFRDVEHGDIVTIGECRPL 131
Query 109 SKTIRFNVLKVESNQ 123
SKT+RFNVLKV Q
Sbjct 132 SKTVRFNVLKVSKGQ 146
> SPAC31G5.03
Length=152
Score = 109 bits (272), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 64/144 (44%), Positives = 83/144 (57%), Gaps = 40/144 (27%)
Query 10 ATQQDVQTEKAFQRQHGASQLALAQSSKKG-GRR---DTGGSM----------------- 48
AT+ VQ+E+AFQ+Q + Q++KKG GRR D G
Sbjct 2 ATELVVQSERAFQKQ-----PHIFQNAKKGAGRRWYKDVGLGFKTPAEAIYGEYVDKKCP 56
Query 49 -------------GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVK 95
G V+S KM R ++IRR YLHF+ KY+R EKRHKN+ H+SP F ++
Sbjct 57 FVGQVSIRGRILTGTVVSTKMHRTIIIRREYLHFIPKYNRYEKRHKNLAAHVSPAF-RIN 115
Query 96 EGDIVTAGQCRPLSKTIRFNVLKV 119
EGD+VT GQCRPLSKT+RFNVL+V
Sbjct 116 EGDVVTVGQCRPLSKTVRFNVLRV 139
> SPAC144.11
Length=152
Score = 109 bits (272), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 64/144 (44%), Positives = 83/144 (57%), Gaps = 40/144 (27%)
Query 10 ATQQDVQTEKAFQRQHGASQLALAQSSKKG-GRR---DTGGSM----------------- 48
AT+ VQ+E+AFQ+Q + Q++KKG GRR D G
Sbjct 2 ATELVVQSERAFQKQ-----PHIFQNAKKGAGRRWYKDVGLGFKTPAEAIYGEYVDKKCP 56
Query 49 -------------GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVK 95
G V+S KM R ++IRR YLHF+ KY+R EKRHKN+ H+SP F ++
Sbjct 57 FVGQVSIRGRILTGTVVSTKMHRTIIIRREYLHFIPKYNRYEKRHKNLAAHVSPAF-RIN 115
Query 96 EGDIVTAGQCRPLSKTIRFNVLKV 119
EGD+VT GQCRPLSKT+RFNVL+V
Sbjct 116 EGDVVTVGQCRPLSKTVRFNVLRV 139
> YDR025w
Length=156
Score = 108 bits (269), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 63/158 (39%), Positives = 84/158 (53%), Gaps = 35/158 (22%)
Query 10 ATQQDVQTEKAFQRQHGASQLALAQSSKKGGRRDTGGSMGM------------------- 50
+T+ VQ+E+AFQ+Q ++SK+ R +G
Sbjct 2 STELTVQSERAFQKQPHIFNNPKVKTSKRTKRWYKNAGLGFKTPKTAIEGSYIDKKCPFT 61
Query 51 -------------VISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEG 97
V+S KM R +VIRR YLH++ KY+R EKRHKNV H+SP F +V+ G
Sbjct 62 GLVSIRGKILTGTVVSTKMHRTIVIRRAYLHYIPKYNRYEKRHKNVPVHVSPAF-RVQVG 120
Query 98 DIVTAGQCRPLSKTIRFNVLKVESNQVFGNVRKQFRLF 135
DIVT GQCRP+SKT+RFNV+KV + G KQF F
Sbjct 121 DIVTVGQCRPISKTVRFNVVKVSAAA--GKANKQFAKF 156
> YBR048w
Length=156
Score = 108 bits (269), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 63/158 (39%), Positives = 84/158 (53%), Gaps = 35/158 (22%)
Query 10 ATQQDVQTEKAFQRQHGASQLALAQSSKKGGRRDTGGSMGM------------------- 50
+T+ VQ+E+AFQ+Q ++SK+ R +G
Sbjct 2 STELTVQSERAFQKQPHIFNNPKVKTSKRTKRWYKNAGLGFKTPKTAIEGSYIDKKCPFT 61
Query 51 -------------VISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEG 97
V+S KM R +VIRR YLH++ KY+R EKRHKNV H+SP F +V+ G
Sbjct 62 GLVSIRGKILTGTVVSTKMHRTIVIRRAYLHYIPKYNRYEKRHKNVPVHVSPAF-RVQVG 120
Query 98 DIVTAGQCRPLSKTIRFNVLKVESNQVFGNVRKQFRLF 135
DIVT GQCRP+SKT+RFNV+KV + G KQF F
Sbjct 121 DIVTVGQCRPISKTVRFNVVKVSAAA--GKANKQFAKF 156
> At4g30800
Length=159
Score = 105 bits (263), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 49/71 (69%), Positives = 58/71 (81%), Gaps = 1/71 (1%)
Query 49 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPL 108
G SAKM+R +++RR YLHFV+KY R EKRH N+ H+SPCF +VKEGD VT GQCRPL
Sbjct 74 GTCHSAKMQRTIIVRRDYLHFVKKYRRYEKRHSNIPAHVSPCF-RVKEGDRVTIGQCRPL 132
Query 109 SKTIRFNVLKV 119
SKT+RFNVLKV
Sbjct 133 SKTVRFNVLKV 143
> CE05860
Length=155
Score = 105 bits (261), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 48/87 (55%), Positives = 61/87 (70%), Gaps = 3/87 (3%)
Query 49 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPL 108
G+V+ KM R +V+RR YLH+++KY R EKRHKNV H SP F + GD+VT G+CRPL
Sbjct 72 GVVLKNKMTRTIVVRRDYLHYIKKYRRYEKRHKNVPAHCSPAFRDIHPGDLVTIGECRPL 131
Query 109 SKTIRFNVLKVESNQVFGNVRKQFRLF 135
SKT+RFNVLKV + G +K F F
Sbjct 132 SKTVRFNVLKVNKS---GTSKKGFSKF 155
> At5g23740
Length=159
Score = 104 bits (260), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 48/71 (67%), Positives = 57/71 (80%), Gaps = 1/71 (1%)
Query 49 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPL 108
G SAKM+R +++RR YLHFV+KY R EKRH N+ H+SPCF +VKEGD V GQCRPL
Sbjct 74 GTCHSAKMQRTIIVRRNYLHFVKKYQRYEKRHSNIPAHVSPCF-RVKEGDHVIIGQCRPL 132
Query 109 SKTIRFNVLKV 119
SKT+RFNVLKV
Sbjct 133 SKTVRFNVLKV 143
> At3g48930
Length=160
Score = 103 bits (258), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 47/71 (66%), Positives = 57/71 (80%), Gaps = 1/71 (1%)
Query 49 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPL 108
G SAKM+R +++RR YLHFV+KY R EKRH N+ H+SPCF +VKEGD + GQCRPL
Sbjct 74 GTCHSAKMQRTIIVRRDYLHFVKKYQRYEKRHSNIPAHVSPCF-RVKEGDHIIIGQCRPL 132
Query 109 SKTIRFNVLKV 119
SKT+RFNVLKV
Sbjct 133 SKTVRFNVLKV 143
> Hs17460599
Length=329
Score = 88.2 bits (217), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 40/71 (56%), Positives = 52/71 (73%), Gaps = 1/71 (1%)
Query 49 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPL 108
GMV KM+R VI R YLH+ KY+ +EK HKN++ +LSPCF ++ DI+T G+C PL
Sbjct 235 GMVTKMKMQR-TVICRDYLHYTCKYNHLEKHHKNMSVNLSPCFRDIQISDIITVGECWPL 293
Query 109 SKTIRFNVLKV 119
SK +RFNVLKV
Sbjct 294 SKMVRFNVLKV 304
> ECU04g0640
Length=156
Score = 63.9 bits (154), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 32/85 (37%), Positives = 49/85 (57%), Gaps = 9/85 (10%)
Query 49 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFE-QVKEGDIVTAGQCRP 107
G+V K ++ +V+ Y+H+ +KY R +RH N + H+SPCFE + GD V G+ RP
Sbjct 70 GVVKKMKAEKTIVVVANYVHYSKKYKRYSRRHTNFSVHMSPCFEGMINVGDTVICGETRP 129
Query 108 LSKTIRFNVLKVESNQVFGNVRKQF 132
LSKT +S+ V G ++K
Sbjct 130 LSKT--------KSSVVLGYIKKAL 146
> Hs22046070
Length=105
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/36 (55%), Positives = 27/36 (75%), Gaps = 3/36 (8%)
Query 98 DIVTAGQCRPLSKTIRFNVLKVESNQVFGNVRKQFR 133
DIVT G+C+PLSKT+RF+VLKV + +KQF+
Sbjct 71 DIVTVGECQPLSKTVRFDVLKVTKA---ASTKKQFQ 103
> Hs22050109
Length=1575
Score = 35.0 bits (79), Expect = 0.043, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 23/43 (53%), Gaps = 0/43 (0%)
Query 71 RKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPLSKTIR 113
+K SR +++ + CHL PC Q KE ++ CR + +R
Sbjct 1285 KKVSRCFRKYTELFCHLDPCLLQSKESQLLQEENCRKKLEALR 1327
> CE26967
Length=1043
Score = 32.0 bits (71), Expect = 0.37, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 91 FEQVKEGDIVTAGQCRPLSKTIRFNVLKVESNQVFGNVRKQFR 133
+ V+E D+++ G RP K +R K+ + +FG V+K F+
Sbjct 42 LQYVEEVDLLSVGMSRPEQKRLRKEYTKMFPSGIFGKVKKAFK 84
> SPBC409.14c
Length=90
Score = 30.8 bits (68), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 38/80 (47%), Gaps = 15/80 (18%)
Query 48 MGMVISAKMKRCVVIRRTYLHF---VRKYSRVEKRH---KNVTCHLSPCFEQVKEGDIVT 101
+G+VIS M + +R F V+KY + + ++ C++ GD VT
Sbjct 6 VGIVISRAMPKTAKVRVAKEKFHPVVKKYVTQYQNYMVQDDLNCNV---------GDAVT 56
Query 102 AGQCRPLSKTIRFNVLKVES 121
CRP S T RF ++K+ S
Sbjct 57 IQPCRPRSATKRFEIIKILS 76
> At3g10270
Length=657
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 19/54 (35%), Positives = 29/54 (53%), Gaps = 3/54 (5%)
Query 36 SKKGGRRDTGGSMGMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSP 89
S GG G S+ +S ++ V++RR + F +KYSR K +TCH+ P
Sbjct 119 SVSGGLHGVGLSVVNALSEALE--VIVRRDGMEFQQKYSR-GKPVTTLTCHVLP 169
> CE13245
Length=1019
Score = 29.3 bits (64), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 11/33 (33%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 41 RRDTGGSMGMVISAKMKRCVVIRRTYLHFVRKY 73
RR+T + ++I + + ++IRR HF KY
Sbjct 304 RRETNNPLRLIIMSATLQVIIIRRKIAHFSTKY 336
> At1g79850
Length=149
Score = 28.5 bits (62), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 31/71 (43%), Gaps = 2/71 (2%)
Query 49 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPL 108
G V+ A + V + L KY R + K H P Q K GD+V + RP+
Sbjct 55 GRVVCATSDKTVAVEVVRLAPHPKYKRRVRMKKKYQAH-DPD-NQFKVGDVVRLEKSRPI 112
Query 109 SKTIRFNVLKV 119
SKT F L V
Sbjct 113 SKTKSFVALPV 123
> At1g49400
Length=116
Score = 28.5 bits (62), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 33/72 (45%), Gaps = 2/72 (2%)
Query 48 MGMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRP 107
+G V+S KM++ VV+ L + Y+R KR H + GD V RP
Sbjct 5 IGTVVSNKMQKSVVVAVDRLFHNKIYNRYVKRTSKFMAHDD--KDACNIGDRVKLDPSRP 62
Query 108 LSKTIRFNVLKV 119
LSK + V ++
Sbjct 63 LSKNKHWIVAEI 74
> CE03855
Length=475
Score = 28.1 bits (61), Expect = 5.4, Method: Composition-based stats.
Identities = 12/61 (19%), Positives = 29/61 (47%), Gaps = 0/61 (0%)
Query 44 TGGSMGMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAG 103
T +GM ++ +K + + R + H++ K + K+ +N + SP + ++ G
Sbjct 207 TIADLGMFLTRFLKNLLTMARRFAHYLVKLYQKAKKQRNKSQKTSPVMPDSERSEVWNTG 266
Query 104 Q 104
+
Sbjct 267 K 267
> YMR188c
Length=237
Score = 27.7 bits (60), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 43/89 (48%), Gaps = 8/89 (8%)
Query 48 MGMVIS-AKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCR 106
+G+V+S KM++ V +R F +K ++ ++ H E +EGD+V R
Sbjct 7 LGLVVSQGKMQKTVKVRVETKVFNKKINKELFHRRDYLVHDEG--EISREGDLVRIEATR 64
Query 107 PLSKTIRFNVLKVESNQVFGNVRKQFRLF 135
PLSK F + ++ N +QF L+
Sbjct 65 PLSKRKFFAI-----AEIIRNKGQQFALY 88
> CE12428
Length=561
Score = 27.7 bits (60), Expect = 7.1, Method: Composition-based stats.
Identities = 15/30 (50%), Positives = 17/30 (56%), Gaps = 1/30 (3%)
Query 69 FVRKYSRVEKRHKNVTCHLSPCFEQVKEGD 98
FV Y KRH N TCH P F+ +KE D
Sbjct 432 FVTVYKGNMKRHLN-TCHPQPEFKSLKEWD 460
Lambda K H
0.322 0.132 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1388795348
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40