bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2500_orf2
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
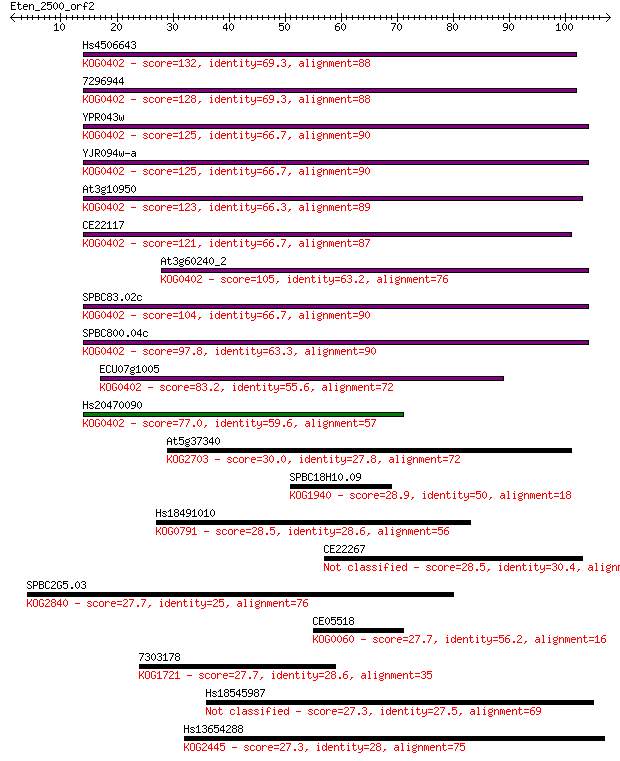
Hs4506643 132 2e-31
7296944 128 3e-30
YPR043w 125 2e-29
YJR094w-a 125 2e-29
At3g10950 123 9e-29
CE22117 121 3e-28
At3g60240_2 105 2e-23
SPBC83.02c 104 4e-23
SPBC800.04c 97.8 5e-21
ECU07g1005 83.2 1e-16
Hs20470090 77.0 8e-15
At5g37340 30.0 1.2
SPBC18H10.09 28.9 2.3
Hs18491010 28.5 2.7
CE22267 28.5 2.9
SPBC2G5.03 27.7 5.5
CE05518 27.7 5.6
7303178 27.7 5.8
Hs18545987 27.3 7.4
Hs13654288 27.3 7.7
> Hs4506643
Length=92
Score = 132 bits (331), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 61/88 (69%), Positives = 74/88 (84%), Gaps = 0/88 (0%)
Query 14 MAKRTKKVGVVGKYGTRYGASLRKVMKKVELQQHAKYTCPFCGKLSVKRQAVGIWHCRGC 73
MAKRTKKVG+VGKYGTRYGASLRK++KK+E+ QHAKYTC FCGK +KR+AVGIWHC C
Sbjct 1 MAKRTKKVGIVGKYGTRYGASLRKMVKKIEISQHAKYTCSFCGKTKMKRRAVGIWHCGSC 60
Query 74 SKTMTGGAWTLQTAAAATVRSTVARLRK 101
KT+ GGAWT T +A TV+S + RL++
Sbjct 61 MKTVAGGAWTYNTTSAVTVKSAIRRLKE 88
> 7296944
Length=92
Score = 128 bits (321), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 61/88 (69%), Positives = 73/88 (82%), Gaps = 0/88 (0%)
Query 14 MAKRTKKVGVVGKYGTRYGASLRKVMKKVELQQHAKYTCPFCGKLSVKRQAVGIWHCRGC 73
MAKRTKKVG+VGKYGTRYGASLRK++KK+E+ QH+KYTC FCGK S+KR VGIW C+ C
Sbjct 1 MAKRTKKVGIVGKYGTRYGASLRKMVKKMEITQHSKYTCSFCGKDSMKRAVVGIWSCKRC 60
Query 74 SKTMTGGAWTLQTAAAATVRSTVARLRK 101
+T+ GGAW T AAA+VRS V RLR+
Sbjct 61 KRTVAGGAWVYSTTAAASVRSAVRRLRE 88
> YPR043w
Length=92
Score = 125 bits (314), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 60/90 (66%), Positives = 74/90 (82%), Gaps = 0/90 (0%)
Query 14 MAKRTKKVGVVGKYGTRYGASLRKVMKKVELQQHAKYTCPFCGKLSVKRQAVGIWHCRGC 73
MAKRTKKVG+ GKYG RYG+SLR+ +KK+E+QQHA+Y C FCGK +VKR A GIW C C
Sbjct 1 MAKRTKKVGITGKYGVRYGSSLRRQVKKLEIQQHARYDCSFCGKKTVKRGAAGIWTCSCC 60
Query 74 SKTMTGGAWTLQTAAAATVRSTVARLRKQM 103
KT+ GGA+T+ TAAAATVRST+ RLR+ +
Sbjct 61 KKTVAGGAYTVSTAAAATVRSTIRRLREMV 90
> YJR094w-a
Length=92
Score = 125 bits (314), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 60/90 (66%), Positives = 74/90 (82%), Gaps = 0/90 (0%)
Query 14 MAKRTKKVGVVGKYGTRYGASLRKVMKKVELQQHAKYTCPFCGKLSVKRQAVGIWHCRGC 73
MAKRTKKVG+ GKYG RYG+SLR+ +KK+E+QQHA+Y C FCGK +VKR A GIW C C
Sbjct 1 MAKRTKKVGITGKYGVRYGSSLRRQVKKLEIQQHARYDCSFCGKKTVKRGAAGIWTCSCC 60
Query 74 SKTMTGGAWTLQTAAAATVRSTVARLRKQM 103
KT+ GGA+T+ TAAAATVRST+ RLR+ +
Sbjct 61 KKTVAGGAYTVSTAAAATVRSTIRRLREMV 90
> At3g10950
Length=92
Score = 123 bits (308), Expect = 9e-29, Method: Compositional matrix adjust.
Identities = 59/89 (66%), Positives = 70/89 (78%), Gaps = 0/89 (0%)
Query 14 MAKRTKKVGVVGKYGTRYGASLRKVMKKVELQQHAKYTCPFCGKLSVKRQAVGIWHCRGC 73
M KRTKK +VGKYGTRYGASLRK +KK+E+ QH KY C FCGK SVKR+ VGIW C+ C
Sbjct 1 MTKRTKKARIVGKYGTRYGASLRKQIKKMEVSQHNKYFCEFCGKYSVKRKVVGIWGCKDC 60
Query 74 SKTMTGGAWTLQTAAAATVRSTVARLRKQ 102
K GGA+T+ TA+A TVRST+ RLR+Q
Sbjct 61 GKVKAGGAYTMNTASAVTVRSTIRRLREQ 89
> CE22117
Length=91
Score = 121 bits (304), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 58/87 (66%), Positives = 71/87 (81%), Gaps = 0/87 (0%)
Query 14 MAKRTKKVGVVGKYGTRYGASLRKVMKKVELQQHAKYTCPFCGKLSVKRQAVGIWHCRGC 73
MAKRTKKVG+VGKYGTRYGASLRK+ KK+E+ QH++YTC FCGK ++KR+A GIW+C C
Sbjct 1 MAKRTKKVGIVGKYGTRYGASLRKMAKKLEVAQHSRYTCSFCGKEAMKRKATGIWNCAKC 60
Query 74 SKTMTGGAWTLQTAAAATVRSTVARLR 100
K + GGA+ T AATVRST+ RLR
Sbjct 61 HKVVAGGAYVYGTVTAATVRSTIRRLR 87
> At3g60240_2
Length=78
Score = 105 bits (262), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 48/76 (63%), Positives = 61/76 (80%), Gaps = 0/76 (0%)
Query 28 GTRYGASLRKVMKKVELQQHAKYTCPFCGKLSVKRQAVGIWHCRGCSKTMTGGAWTLQTA 87
GTRYGAS+RK +KK+E+ QH+KY C FCGK VKR+AVGIW C+ C K GGA+T+ TA
Sbjct 1 GTRYGASIRKQIKKMEVSQHSKYFCEFCGKYGVKRKAVGIWGCKDCGKVKAGGAYTMNTA 60
Query 88 AAATVRSTVARLRKQM 103
+A TVRST+ RLR+Q+
Sbjct 61 SAVTVRSTIRRLREQI 76
> SPBC83.02c
Length=94
Score = 104 bits (260), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 60/92 (65%), Positives = 76/92 (82%), Gaps = 2/92 (2%)
Query 14 MAKRTKKVGVVGKYGTRYGASLRKVMKKVELQQHAKYTCPFCGKLSVKRQAVGIWHC--R 71
M KRTKKVGV GKYG RYGASLR+ ++K+E+QQH++Y CPFCG+L+VKR A GIW C +
Sbjct 1 MTKRTKKVGVTGKYGVRYGASLRRDVRKIEVQQHSRYQCPFCGRLTVKRTAAGIWKCSGK 60
Query 72 GCSKTMTGGAWTLQTAAAATVRSTVARLRKQM 103
GCSKT+ GGAWT+ TAAA + RST+ RLR+ +
Sbjct 61 GCSKTLAGGAWTVTTAAATSARSTIRRLREMV 92
> SPBC800.04c
Length=94
Score = 97.8 bits (242), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 57/92 (61%), Positives = 73/92 (79%), Gaps = 2/92 (2%)
Query 14 MAKRTKKVGVVGKYGTRYGASLRKVMKKVELQQHAKYTCPFCGKLSVKRQAVGIWHC--R 71
M KRTKKVGV GKYG RYGASLR+ ++K+E+QQH++Y CPFCG+ +VKR A GIW C +
Sbjct 1 MTKRTKKVGVTGKYGVRYGASLRRDVRKIEVQQHSRYQCPFCGRNTVKRTAAGIWCCNGK 60
Query 72 GCSKTMTGGAWTLQTAAAATVRSTVARLRKQM 103
GC K + GGAWT+ TAAA + RST+ RLR+ +
Sbjct 61 GCKKVLAGGAWTVTTAAATSARSTIRRLREMV 92
> ECU07g1005
Length=88
Score = 83.2 bits (204), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 40/72 (55%), Positives = 49/72 (68%), Gaps = 0/72 (0%)
Query 17 RTKKVGVVGKYGTRYGASLRKVMKKVELQQHAKYTCPFCGKLSVKRQAVGIWHCRGCSKT 76
RTKKVG+ G +G RYG++LRK MK V Q AKY C CGK +VKR+AVGIW CR C +
Sbjct 3 RTKKVGIAGSFGARYGSTLRKRMKAVLETQRAKYICEACGKKTVKREAVGIWKCRSCRRV 62
Query 77 MTGGAWTLQTAA 88
GG++ T A
Sbjct 63 FAGGSYAKTTPA 74
> Hs20470090
Length=93
Score = 77.0 bits (188), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 34/57 (59%), Positives = 44/57 (77%), Gaps = 0/57 (0%)
Query 14 MAKRTKKVGVVGKYGTRYGASLRKVMKKVELQQHAKYTCPFCGKLSVKRQAVGIWHC 70
M K KKV ++ KYGT YGASL+K++KK+ + QHA+ TC FCGK+ +KRQAVG W C
Sbjct 1 MIKANKKVRIINKYGTHYGASLQKMVKKIIIHQHAEDTCSFCGKIRIKRQAVGTWCC 57
> At5g37340
Length=600
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 20/72 (27%), Positives = 35/72 (48%), Gaps = 3/72 (4%)
Query 29 TRYGASLRKVMKKVELQQHAKYTCPFCGKLSVKRQAVGIWHCRGCSKTMTGGAWTLQTAA 88
TR+ +L +KV + + + CP CG+ + + Q G RGCS + A ++T
Sbjct 96 TRFLLTLIPHFRKVLI---SAFECPHCGERNNEVQFAGEIQPRGCSYHLEVSAGDVKTFD 152
Query 89 AATVRSTVARLR 100
V+S A ++
Sbjct 153 RQVVKSESATIK 164
> SPBC18H10.09
Length=428
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 9/18 (50%), Positives = 12/18 (66%), Gaps = 0/18 (0%)
Query 51 TCPFCGKLSVKRQAVGIW 68
TCP CG ++VK+Q W
Sbjct 384 TCPHCGNMTVKKQTSAYW 401
> Hs18491010
Length=1997
Score = 28.5 bits (62), Expect = 2.7, Method: Composition-based stats.
Identities = 16/56 (28%), Positives = 23/56 (41%), Gaps = 0/56 (0%)
Query 27 YGTRYGASLRKVMKKVELQQHAKYTCPFCGKLSVKRQAVGIWHCRGCSKTMTGGAW 82
YG R G S + +K V Y+ P G + K + HCR + T +W
Sbjct 1320 YGLRPGRSYQFNVKTVSGDSWKTYSKPIFGSVRTKPDKIQNLHCRPQNSTAIACSW 1375
> CE22267
Length=301
Score = 28.5 bits (62), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 28/47 (59%), Gaps = 1/47 (2%)
Query 57 KLSVKRQAVGIWHCRGCSKTMTGGAWTLQTAAAATVRSTVA-RLRKQ 102
K++ K + +G++ C K + GAW++ T TV++++ RLR +
Sbjct 38 KIARKNENIGVYLCCQRLKDIGEGAWSVDTEYRLTVKNSIGKRLRNK 84
> SPBC2G5.03
Length=335
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 19/76 (25%), Positives = 30/76 (39%), Gaps = 0/76 (0%)
Query 4 PFRFRGEIKKMAKRTKKVGVVGKYGTRYGASLRKVMKKVELQQHAKYTCPFCGKLSVKRQ 63
P FRG + M K+ + + Y ++ V+ Q + TC CG +S R
Sbjct 239 PEAFRGTARAMIKQLENIRPSSILDIIYSGESMQLASSVQEQLPQQTTCERCGFISSNRI 298
Query 64 AVGIWHCRGCSKTMTG 79
G +K +TG
Sbjct 299 CKACMLLEGLNKGITG 314
> CE05518
Length=660
Score = 27.7 bits (60), Expect = 5.6, Method: Composition-based stats.
Identities = 9/16 (56%), Positives = 11/16 (68%), Gaps = 0/16 (0%)
Query 55 CGKLSVKRQAVGIWHC 70
CGK S+ R G+WHC
Sbjct 461 CGKSSLLRMFAGLWHC 476
> 7303178
Length=1290
Score = 27.7 bits (60), Expect = 5.8, Method: Composition-based stats.
Identities = 10/35 (28%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 24 VGKYGTRYGASLRKVMKKVELQQHAKYTCPFCGKL 58
+G YG+ +G S ++ + +H + C +C KL
Sbjct 218 MGWYGSEFGLSAYGMLMAMSHAEHLPFKCEYCSKL 252
> Hs18545987
Length=303
Score = 27.3 bits (59), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 30/73 (41%), Gaps = 8/73 (10%)
Query 36 RKVMKKVELQQHAKYTCPFCGKLSVKRQAVGIWHCRGCSKTMTGGAWTLQTAAAA----T 91
R ++ + H CP L KR +G W C GC K +T ++ T A
Sbjct 13 RPILSFHYFRLHRGEVCP----LWHKRVCLGKWPCTGCQKRLTDPGVSMHTIEADLALYL 68
Query 92 VRSTVARLRKQMG 104
V + + R + +G
Sbjct 69 VYTKLTRFKSGLG 81
> Hs13654288
Length=360
Score = 27.3 bits (59), Expect = 7.7, Method: Composition-based stats.
Identities = 21/86 (24%), Positives = 32/86 (37%), Gaps = 11/86 (12%)
Query 32 GASLRKVMKKVELQQHAKYTCPFCGKLSVK---RQAVGIWHCRGCSKTMTGGAWTL---- 84
A + ++ V H + SVK + G WHC KT +G W +
Sbjct 8 AADHKDLIHDVSFDFHGRRMATCSSDQSVKVWDKSESGDWHCTASWKTHSGSVWRVTWAH 67
Query 85 ----QTAAAATVRSTVARLRKQMGES 106
Q A+ + T A + +GES
Sbjct 68 PEFGQVLASCSFDRTAAVWEEIVGES 93
Lambda K H
0.322 0.131 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164169380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40