bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2540_orf1
Length=162
Score E
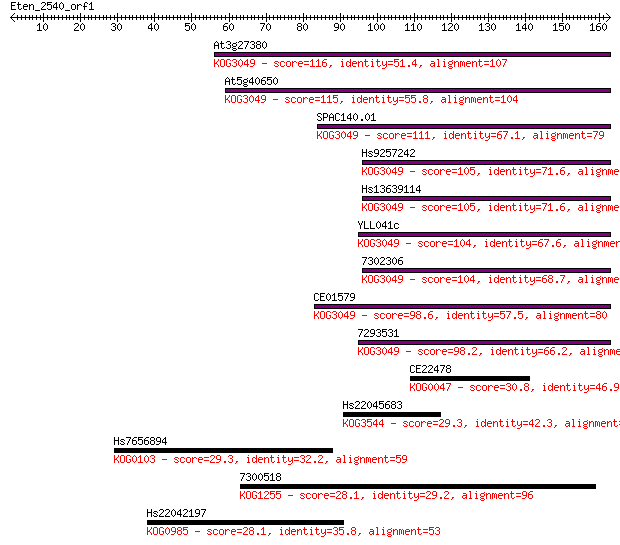
Sequences producing significant alignments: (Bits) Value
At3g27380 116 2e-26
At5g40650 115 3e-26
SPAC140.01 111 6e-25
Hs9257242 105 3e-23
Hs13639114 105 3e-23
YLL041c 104 7e-23
7302306 104 8e-23
CE01579 98.6 4e-21
7293531 98.2 6e-21
CE22478 30.8 1.1
Hs22045683 29.3 3.4
Hs7656894 29.3 3.7
7300518 28.1 6.9
Hs22042197 28.1 7.2
> At3g27380
Length=279
Score = 116 bits (290), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 55/107 (51%), Positives = 74/107 (69%), Gaps = 2/107 (1%)
Query 56 PAGVSAACRLLFPRASPVLSGLVSQQRASFSSNAGGTNLTFRIHRYDPEKGGRPHMQEYT 115
P+ ++ A RL+ R + +G ++ +AS G TF+I+R++P+ G+P +Q Y
Sbjct 14 PSKLATAARLIPARWTS--TGAEAETKASSGGGRGSNLKTFQIYRWNPDNPGKPELQNYQ 71
Query 116 LDTSTCGPMVLDALIAIKDRQDPTLTFRRSCREGICGSCAMNIDGIN 162
+D CGPMVLDALI IK+ DP+LTFRRSCREGICGSCAMNIDG N
Sbjct 72 IDLKDCGPMVLDALIKIKNEMDPSLTFRRSCREGICGSCAMNIDGCN 118
> At5g40650
Length=280
Score = 115 bits (289), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 58/105 (55%), Positives = 77/105 (73%), Gaps = 5/105 (4%)
Query 59 VSAACRLLFPRASPVLSGLVSQQRASFSSNAGGTNL-TFRIHRYDPEKGGRPHMQEYTLD 117
+S A RL+ R + +G +Q +AS + GG +L TF+I+R++P+ G+P +Q+Y +D
Sbjct 17 LSTAARLIPARWTS--TGSEAQSKAS--TGGGGASLKTFQIYRWNPDNPGKPELQDYKID 72
Query 118 TSTCGPMVLDALIAIKDRQDPTLTFRRSCREGICGSCAMNIDGIN 162
CGPMVLDALI IK+ DP+LTFRRSCREGICGSCAMNIDG N
Sbjct 73 LKDCGPMVLDALIKIKNEMDPSLTFRRSCREGICGSCAMNIDGCN 117
> SPAC140.01
Length=252
Score = 111 bits (277), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 53/81 (65%), Positives = 65/81 (80%), Gaps = 2/81 (2%)
Query 84 SFSSNAGGTNL-TFRIHRYDPEKGG-RPHMQEYTLDTSTCGPMVLDALIAIKDRQDPTLT 141
S + + G NL TF I+R++PEK +P +Q+YT+D + CGPMVLDALI IK+ QDPTLT
Sbjct 11 SANPQSQGENLKTFEIYRWNPEKPEVKPKLQKYTVDLTKCGPMVLDALIKIKNEQDPTLT 70
Query 142 FRRSCREGICGSCAMNIDGIN 162
FRRSCREGICGSCAMNI+G N
Sbjct 71 FRRSCREGICGSCAMNINGSN 91
> Hs9257242
Length=280
Score = 105 bits (263), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 48/68 (70%), Positives = 56/68 (82%), Gaps = 1/68 (1%)
Query 96 FRIHRYDPEKGG-RPHMQEYTLDTSTCGPMVLDALIAIKDRQDPTLTFRRSCREGICGSC 154
F I+R+DP+K G +PHMQ Y +D + CGPMVLDALI IK+ D TLTFRRSCREGICGSC
Sbjct 42 FAIYRWDPDKAGDKPHMQTYKVDLNKCGPMVLDALIKIKNEVDSTLTFRRSCREGICGSC 101
Query 155 AMNIDGIN 162
AMNI+G N
Sbjct 102 AMNINGGN 109
> Hs13639114
Length=280
Score = 105 bits (263), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 48/68 (70%), Positives = 56/68 (82%), Gaps = 1/68 (1%)
Query 96 FRIHRYDPEKGG-RPHMQEYTLDTSTCGPMVLDALIAIKDRQDPTLTFRRSCREGICGSC 154
F I+R+DP+K G +PHMQ Y +D + CGPMVLDALI IK+ D TLTFRRSCREGICGSC
Sbjct 42 FAIYRWDPDKAGDKPHMQTYEVDLNKCGPMVLDALIKIKNEVDSTLTFRRSCREGICGSC 101
Query 155 AMNIDGIN 162
AMNI+G N
Sbjct 102 AMNINGGN 109
> YLL041c
Length=266
Score = 104 bits (260), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 46/69 (66%), Positives = 56/69 (81%), Gaps = 1/69 (1%)
Query 95 TFRIHRYDP-EKGGRPHMQEYTLDTSTCGPMVLDALIAIKDRQDPTLTFRRSCREGICGS 153
TF+++R++P E +PH+Q Y +D + CGPMVLDAL+ IKD QD TLTFRRSCREGICGS
Sbjct 35 TFKVYRWNPDEPSAKPHLQSYQVDLNDCGPMVLDALLKIKDEQDSTLTFRRSCREGICGS 94
Query 154 CAMNIDGIN 162
CAMNI G N
Sbjct 95 CAMNIGGRN 103
> 7302306
Length=297
Score = 104 bits (259), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 46/68 (67%), Positives = 54/68 (79%), Gaps = 1/68 (1%)
Query 96 FRIHRYDPEKGG-RPHMQEYTLDTSTCGPMVLDALIAIKDRQDPTLTFRRSCREGICGSC 154
F I+R++P+ G +P+MQ Y +D CGPMVLDALI IK+ DPTLTFRRSCREGICGSC
Sbjct 49 FEIYRWNPDNAGEKPYMQTYEVDLRECGPMVLDALIKIKNEMDPTLTFRRSCREGICGSC 108
Query 155 AMNIDGIN 162
AMNI G N
Sbjct 109 AMNIGGTN 116
> CE01579
Length=298
Score = 98.6 bits (244), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 46/81 (56%), Positives = 58/81 (71%), Gaps = 1/81 (1%)
Query 83 ASFSSNAGGTNLTFRIHRYDPEK-GGRPHMQEYTLDTSTCGPMVLDALIAIKDRQDPTLT 141
A+ + G TF I+R++PE G +P +Q++ +D CG M+LDALI IK+ DPTLT
Sbjct 43 AAKTKKTGNRIKTFEIYRFNPEAPGAKPTVQKFDVDLDQCGTMILDALIKIKNEVDPTLT 102
Query 142 FRRSCREGICGSCAMNIDGIN 162
FRRSCREGICGSCAMNI G N
Sbjct 103 FRRSCREGICGSCAMNIGGQN 123
> 7293531
Length=398
Score = 98.2 bits (243), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 45/68 (66%), Positives = 51/68 (75%), Gaps = 2/68 (2%)
Query 95 TFRIHRYDPEKGGRPHMQEYTLDTSTCGPMVLDALIAIKDRQDPTLTFRRSCREGICGSC 154
TF I+R+ P G +P Q Y +D CG MVLDALI IK+ DPTLTFRRSCREGICGSC
Sbjct 150 TFEIYRWKP--GDQPQTQTYEVDLEQCGAMVLDALIKIKNEMDPTLTFRRSCREGICGSC 207
Query 155 AMNIDGIN 162
AMNI+G N
Sbjct 208 AMNINGTN 215
> CE22478
Length=512
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 20/33 (60%), Gaps = 1/33 (3%)
Query 109 PHMQEYTLDTSTCGPMVLDALIAIKDR-QDPTL 140
PHM T++ + GPM D L A +DR Q+P L
Sbjct 5 PHMVNLTVNDTKPGPMAEDQLKAYRDRNQEPHL 37
> Hs22045683
Length=1350
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 11/26 (42%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 91 GTNLTFRIHRYDPEKGGRPHMQEYTL 116
G L F + +D KGGRP +Q+Y +
Sbjct 1048 GKALNFTLPFFDSSKGGRPSVQQYLI 1073
> Hs7656894
Length=839
Score = 29.3 bits (64), Expect = 3.7, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 27/62 (43%), Gaps = 3/62 (4%)
Query 29 LSEAQPSEWPVNSLIFMEMRRLTSSLSPAGVSAACRLLFPRASPVLSGLVSQ---QRASF 85
L A S+ P+N FM ++S ++ A C L R P L ++ Q QR
Sbjct 276 LMSANASDLPLNIECFMNDLDVSSKMNRAQFEQLCASLLARVEPPLKAVMEQANLQREDI 335
Query 86 SS 87
SS
Sbjct 336 SS 337
> 7300518
Length=342
Score = 28.1 bits (61), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 28/101 (27%), Positives = 46/101 (45%), Gaps = 16/101 (15%)
Query 63 CRLLFPRASPVLSGLVSQQRASFSSNAG---GTNLTFRIHRYDPEKGGRPHMQEYTLDTS 119
C L +A+ VL + ++A+F S GTN+ + +P+KGG H+ + +
Sbjct 31 CNLKINKATKVLVQGFTGKQATFHSEESIKYGTNI---VGGVNPKKGGTEHLGKPVFKS- 86
Query 120 TCGPMVLDALIAIKDRQDPTLTF--RRSCREGICGSCAMNI 158
V +A+ K + D T+ F S EGIC + I
Sbjct 87 -----VAEAV--EKAKPDATVIFIPPPSAAEGICAAIESEI 120
> Hs22042197
Length=586
Score = 28.1 bits (61), Expect = 7.2, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 29/59 (49%), Gaps = 6/59 (10%)
Query 38 PVNSLIFMEMRRLTSSLSPAGVSAACRL------LFPRASPVLSGLVSQQRASFSSNAG 90
P+ L+F E +TS P V AA L L + +++ V+Q+R +FS AG
Sbjct 344 PLPLLLFFEALFITSHAFPCPVDAALTLEGIKCGLSEKRLDLVTNWVTQERLTFSEEAG 402
Lambda K H
0.320 0.132 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2244926132
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40