bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2552_orf1
Length=197
Score E
Sequences producing significant alignments: (Bits) Value
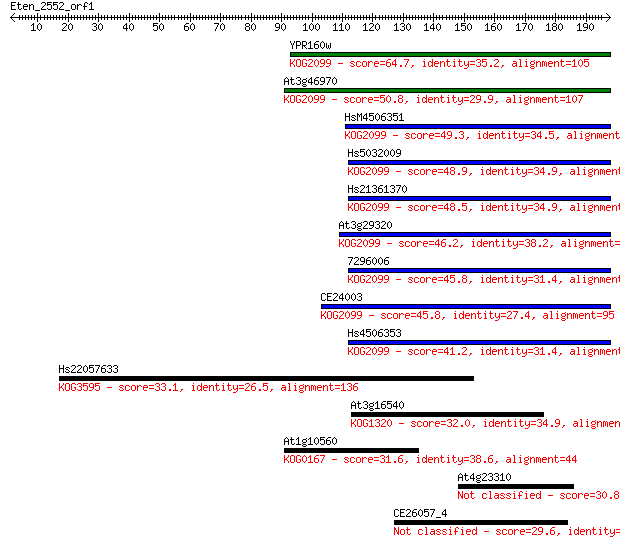
YPR160w 64.7 1e-10
At3g46970 50.8 2e-06
HsM4506351 49.3 5e-06
Hs5032009 48.9 6e-06
Hs21361370 48.5 8e-06
At3g29320 46.2 5e-05
7296006 45.8 5e-05
CE24003 45.8 6e-05
Hs4506353 41.2 0.001
Hs22057633 33.1 0.41
At3g16540 32.0 0.84
At1g10560 31.6 1.1
At4g23310 30.8 2.0
CE26057_4 29.6 3.6
> YPR160w
Length=902
Score = 64.7 bits (156), Expect = 1e-10, Method: Composition-based stats.
Identities = 37/105 (35%), Positives = 53/105 (50%), Gaps = 5/105 (4%)
Query 93 KLTGAVPRTIPGMYNVVEDPHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTR 152
+LTG +P+ I + D K LW + + Q ++HVE TLA++
Sbjct 29 RLTGFLPQEIKSI-----DTMIPLKSRALWNKHQVKKFNKAEDFQDRFIDHVETTLARSL 83
Query 153 FNLDPESCYRAAAFSVRDRLIETLNDTNAFFHEKDVKRAYYLSLE 197
+N D + Y AA+ S+RD L+ N T F +D KR YYLSLE
Sbjct 84 YNCDDMAAYEAASMSIRDNLVIDWNKTQQKFTTRDPKRVYYLSLE 128
> At3g46970
Length=841
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 32/107 (29%), Positives = 48/107 (44%), Gaps = 16/107 (14%)
Query 91 FSKLTGAVPRTIPGMYNVVEDPHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAK 150
+ G ++P + +P ADD E I +IV H +Y+
Sbjct 1 MANANGKAATSLPEKISAKANPEADDATE----------------IAGNIVYHAKYSPHF 44
Query 151 TRFNLDPESCYRAAAFSVRDRLIETLNDTNAFFHEKDVKRAYYLSLE 197
+ PE A A S+RDRLI+ N+T F++ D K+ YYLS+E
Sbjct 45 SPLKFGPEQALYATAESLRDRLIQLWNETYVHFNKVDPKQTYYLSME 91
> HsM4506351
Length=843
Score = 49.3 bits (116), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 30/87 (34%), Positives = 44/87 (50%), Gaps = 0/87 (0%)
Query 111 DPHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRD 170
+P D +K K + DV +++S H+ +TL K R P + A A +VRD
Sbjct 3 EPLTDSEKRKQISVRGLAGLGDVAEVRKSFNRHLHFTLVKDRNVATPRDYFFALAHTVRD 62
Query 171 RLIETLNDTNAFFHEKDVKRAYYLSLE 197
L+ T ++E+D KR YYLSLE
Sbjct 63 HLVGRWIRTQQHYYERDPKRIYYLSLE 89
> Hs5032009
Length=842
Score = 48.9 bits (115), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 30/86 (34%), Positives = 44/86 (51%), Gaps = 0/86 (0%)
Query 112 PHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDR 171
P +D +K K + +V ++++ H+ +TL K R P Y A A +VRD
Sbjct 4 PLSDQEKRKQISVRGLAGVENVTELKKNFNRHLHFTLVKDRNVATPRDYYFALAHTVRDH 63
Query 172 LIETLNDTNAFFHEKDVKRAYYLSLE 197
L+ T ++EKD KR YYLSLE
Sbjct 64 LVGRWIRTQQHYYEKDPKRIYYLSLE 89
> Hs21361370
Length=843
Score = 48.5 bits (114), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 30/86 (34%), Positives = 43/86 (50%), Gaps = 0/86 (0%)
Query 112 PHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDR 171
P D +K K + DV +++S H+ +TL K R P + A A +VRD
Sbjct 4 PLTDSEKRKQISVRGLAGLGDVAEVRKSFNRHLHFTLVKDRNVATPRDYFFALAHTVRDH 63
Query 172 LIETLNDTNAFFHEKDVKRAYYLSLE 197
L+ T ++E+D KR YYLSLE
Sbjct 64 LVGRWIRTQQHYYERDPKRIYYLSLE 89
> At3g29320
Length=962
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 34/91 (37%), Positives = 50/91 (54%), Gaps = 5/91 (5%)
Query 109 VEDPHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYT--LAKTRFNLDPESCYRAAAF 166
V D D ++E M + P D S+ SI H E+T + +F L P++ + A A
Sbjct 70 VTDAVLDSEQEVFISSMNPFAP-DAASVASSIKYHAEFTPLFSPEKFEL-PKAFF-ATAQ 126
Query 167 SVRDRLIETLNDTNAFFHEKDVKRAYYLSLE 197
SVRD LI N T +++ +VK+AYYLS+E
Sbjct 127 SVRDALIMNWNATYEYYNRVNVKQAYYLSME 157
> 7296006
Length=844
Score = 45.8 bits (107), Expect = 5e-05, Method: Composition-based stats.
Identities = 27/86 (31%), Positives = 42/86 (48%), Gaps = 0/86 (0%)
Query 112 PHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDR 171
P +D + K + +V ++++ H+ YTL K R Y A A +V+D
Sbjct 4 PQSDADRRKQISVRGIAEVGNVTEVKKNFNRHLHYTLVKDRNVSTLRDYYFALANTVKDN 63
Query 172 LIETLNDTNAFFHEKDVKRAYYLSLE 197
++ T ++EKD KR YYLSLE
Sbjct 64 MVGRWIRTQQHYYEKDPKRVYYLSLE 89
> CE24003
Length=882
Score = 45.8 bits (107), Expect = 6e-05, Method: Composition-based stats.
Identities = 26/95 (27%), Positives = 50/95 (52%), Gaps = 3/95 (3%)
Query 103 PGMYNVVEDPHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTRFNLDPESCYR 162
P +++++ + H K+ + + + +V +I+++ H+ +++ K R Y
Sbjct 32 PKLFSMITNDHDRRKQISVRGIAQV---ENVSNIKKAFNRHLHFSIIKDRNVATDRDYYF 88
Query 163 AAAFSVRDRLIETLNDTNAFFHEKDVKRAYYLSLE 197
A A +VRD L+ T +++KD KR YYLSLE
Sbjct 89 ALANTVRDHLVSRWIRTQQHYYDKDPKRVYYLSLE 123
> Hs4506353
Length=847
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 41/86 (47%), Gaps = 0/86 (0%)
Query 112 PHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDR 171
P D +K + + +V +++S H+ +TL K R Y A A +VRD
Sbjct 4 PLTDQEKRRQISIRGIVGVENVAELKKSFNRHLHFTLVKDRNVATTRDYYFALAHTVRDH 63
Query 172 LIETLNDTNAFFHEKDVKRAYYLSLE 197
L+ T +++K KR YYLSLE
Sbjct 64 LVGRWIRTQQHYYDKCPKRVYYLSLE 89
> Hs22057633
Length=1770
Score = 33.1 bits (74), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 36/142 (25%), Positives = 57/142 (40%), Gaps = 20/142 (14%)
Query 17 LPRPLLRSVSVL--LLPRLEEGAVRLRRTRSAYFLAFFLLFLYLRLLTD----FVKMATG 70
+P+ L V+ L +L L EG + YFL L LL D F +
Sbjct 766 VPQTDLNMVTQLAKMLDALLEGEIEDLDLLECYFLEALYCSLGASLLEDGRMKFDEYIKR 825
Query 71 VEDVFTSDTNQHWEMKRKASFSKLTGAVPRTIPGMYNVVEDPHADDKKEKLWKLMETYLP 130
+ + T DT W A P +PG + D H D+K+ + W +P
Sbjct 826 LASLSTVDTEGVW-------------ANPGELPGQLPTLYDFHFDNKRNQ-WVPWSKLVP 871
Query 131 SDVHSIQRSIVNHVEYTLAKTR 152
+H+ +R +N + +T+ TR
Sbjct 872 EYIHAPERKFINILVHTVDTTR 893
> At3g16540
Length=555
Score = 32.0 bits (71), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 33/66 (50%), Gaps = 4/66 (6%)
Query 113 HADDKKEKLWKLMETYLPSD---VHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVR 169
H D+KK + WK +E P D + S+ + N EY+ +K LD +S R F++
Sbjct 83 HEDEKKLERWKKIEESHPLDELVLDSVVKVFSNSTEYSKSKPWKTLDQKSS-RGTGFAIA 141
Query 170 DRLIET 175
R I T
Sbjct 142 GRKILT 147
> At1g10560
Length=697
Score = 31.6 bits (70), Expect = 1.1, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 26/46 (56%), Gaps = 2/46 (4%)
Query 91 FSKLTGAVPRTIPGMYNVVE-DPHADD-KKEKLWKLMETYLPSDVH 134
+S+L G P IPG+ N+V+ D + D K+ L +M + SD H
Sbjct 504 YSRLIGENPDAIPGLMNIVKGDDYGDSAKRSALLAVMGLLMQSDNH 549
> At4g23310
Length=830
Score = 30.8 bits (68), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 21/40 (52%), Gaps = 2/40 (5%)
Query 148 LAKTRFNLDPESCYRAAAFSVRDRLIETLN--DTNAFFHE 185
L R +L PE C+ AF+V+D LI N D F+ E
Sbjct 84 LFDCRGDLPPEVCHNCVAFAVKDTLIRCPNERDVTLFYDE 123
> CE26057_4
Length=336
Score = 29.6 bits (65), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 25/57 (43%), Gaps = 3/57 (5%)
Query 127 TYLPSDVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDRLIETLNDTNAFF 183
TYL D +SIQ +NH+ Y L+ R N D Y + +D + D N
Sbjct 10 TYLQIDQNSIQ---INHILYRLSVIRSNGDGGRPYDVDEYGYQDYALNVHEDPNEIL 63
Lambda K H
0.325 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3371754244
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40