bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2631_orf1
Length=156
Score E
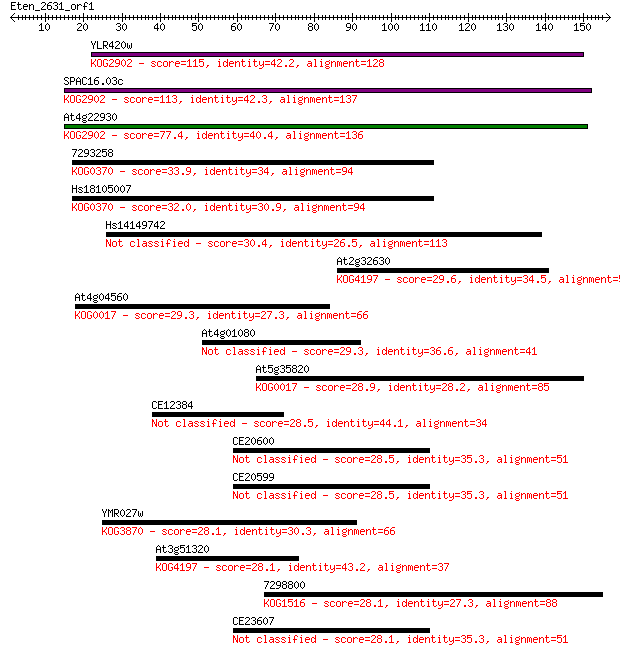
Sequences producing significant alignments: (Bits) Value
YLR420w 115 5e-26
SPAC16.03c 113 2e-25
At4g22930 77.4 1e-14
7293258 33.9 0.15
Hs18105007 32.0 0.44
Hs14149742 30.4 1.6
At2g32630 29.6 2.5
At4g04560 29.3 3.2
At4g01080 29.3 3.3
At5g35820 28.9 4.5
CE12384 28.5 5.0
CE20600 28.5 5.9
CE20599 28.5 6.2
YMR027w 28.1 6.4
At3g51320 28.1 6.4
7298800 28.1 6.8
CE23607 28.1 7.5
> YLR420w
Length=364
Score = 115 bits (287), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 54/130 (41%), Positives = 81/130 (62%), Gaps = 2/130 (1%)
Query 22 DMHTHLRQVELMDMVTLQIRKGGCDRVLVMPNTVPPIVTCSQALEYRNELLKRDPNVNYL 81
DMH H+R+ + ++VT +IR GG +MPN PPI T + +EY+ L K P +L
Sbjct 12 DMHVHVREGAMCELVTPKIRDGGVSIAYIMPNLQPPITTLDRVIEYKKTLQKLAPKTTFL 71
Query 82 MTLYLCREIDADDIAKRAKESHVVGVKLYPRGVTTNSEAGVE--DLTQYEGVFKVMEELG 139
M+ YL +++ D I + A++ + GVK YP GVTTNS AGV+ D + + +FK M+E
Sbjct 72 MSFYLSKDLTPDLIHEAAQQHAIRGVKCYPAGVTTNSAAGVDPNDFSAFYPIFKAMQEEN 131
Query 140 MSLHIHAEKP 149
+ L++H EKP
Sbjct 132 LVLNLHGEKP 141
> SPAC16.03c
Length=337
Score = 113 bits (282), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 58/137 (42%), Positives = 79/137 (57%), Gaps = 1/137 (0%)
Query 15 LVMPMADDMHTHLRQVELMDMVTLQIRKGGCDRVLVMPNTVPPIVTCSQALEYRNELLKR 74
L +P DMH HLRQ ++ V + +GG VMPN +PPI T L+Y+ E+ +
Sbjct 3 LKIPGLADMHVHLRQDNMLKAVVPTVAEGGVSVAYVMPNLIPPITTVDACLQYKKEIEQL 62
Query 75 DPNVNYLMTLYLCREIDADDIAKRAKESHVVGVKLYPRGVTTNSEAGVEDLTQYEGVFKV 134
D YLM+LYL E + I + AK+ + GVK YP+G TTNSE+GVE + F
Sbjct 63 DSKTTYLMSLYLSPETTPEVIYEAAKKG-IRGVKSYPKGATTNSESGVESYEPFYPTFAA 121
Query 135 MEELGMSLHIHAEKPDA 151
M+E GM L+IH E P +
Sbjct 122 MQETGMILNIHGEVPPS 138
> At4g22930
Length=377
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 55/143 (38%), Positives = 77/143 (53%), Gaps = 11/143 (7%)
Query 15 LVMPMADDMHTHLRQVELMDMVTLQIRKGGCDRVLVMPNTVPPIVTCSQALEYRNELLKR 74
L + DD H HLR +L+ V R +VMPN PP+ + + A+ YR ++K
Sbjct 35 LTITQPDDWHLHLRDGDLLHAVVPH-SASNFKRAIVMPNLKPPVTSTAAAIIYRKFIMKA 93
Query 75 DPN---VNYLMTLYLCREIDADDIAKRAKESHVV-GVKLYPRGVTTNSEAGVEDLTQYEG 130
P+ + LMTLYL + ++I + A+ES VV VKLYP G TTNS+ GV DL +
Sbjct 94 LPSESSFDPLMTLYLTDKTLPEEI-RLARESGVVYAVKLYPAGATTNSQDGVTDL--FGK 150
Query 131 VFKVMEEL---GMSLHIHAEKPD 150
V+EE+ M L +H E D
Sbjct 151 CLPVLEEMVKQNMPLLVHGEVTD 173
> 7293258
Length=2189
Score = 33.9 bits (76), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 32/101 (31%), Positives = 44/101 (43%), Gaps = 13/101 (12%)
Query 17 MPMADDMHTHLRQ-----VELMDMVTLQIRKGGCDRVLVMPNTVPPIVTCSQALEYRNEL 71
+P D+H HLR+ E T GG V MPNT P IV +++ EL
Sbjct 1444 LPGFIDVHVHLREPGATHKEDFASGTAAALAGGVTLVCAMPNTNPSIVDRETFTQFQ-EL 1502
Query 72 LKRDPNVNYLMTLYLCREIDADDIAKRAKE--SHVVGVKLY 110
K +Y LY+ +DD + E SH G+K+Y
Sbjct 1503 AKAGARCDY--ALYVGA---SDDNWAQVNELASHACGLKMY 1538
> Hs18105007
Length=2225
Score = 32.0 bits (71), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 29/99 (29%), Positives = 43/99 (43%), Gaps = 9/99 (9%)
Query 17 MPMADDMHTHLRQ-----VELMDMVTLQIRKGGCDRVLVMPNTVPPIVTCSQALEYRNEL 71
+P D+H HLR+ E T GG V MPNT PPI+ + AL +L
Sbjct 1464 LPGLIDVHVHLREPGGTHKEDFASGTAAALAGGITMVCAMPNTRPPIID-APALALAQKL 1522
Query 72 LKRDPNVNYLMTLYLCREIDADDIAKRAKESHVVGVKLY 110
+ ++ + L E +A + A + G+KLY
Sbjct 1523 AEAGARCDFALFLGASSE-NAGTLGTVAGSA--AGLKLY 1558
> Hs14149742
Length=691
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 30/114 (26%), Positives = 49/114 (42%), Gaps = 19/114 (16%)
Query 26 HLRQVELMD-MVTLQIRKGGCDRVLVMPNTVPPIVTCSQALEYRNELLKRDPNVNYLMTL 84
R+ MD +VTL+ GG D +LV+P +A +N+L + N LM L
Sbjct 117 QFREPRPMDELVTLEEADGGSDILLVVP----------KATVLQNQLDESQQERNDLMQL 166
Query 85 YLCREIDADDIAKRAKESHVVGVKLYPRGVTTNSEAGVEDLTQYEGVFKVMEEL 138
L E ++ R +E R + T + E + QY+G+ + E+
Sbjct 167 KLQLEGQVTELRSRVQE--------LERALATARQEHTELMEQYKGISRSHGEI 212
> At2g32630
Length=624
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 19/57 (33%), Positives = 27/57 (47%), Gaps = 2/57 (3%)
Query 86 LCREIDADDIAKRAKESHVVGVK--LYPRGVTTNSEAGVEDLTQYEGVFKVMEELGM 140
LCR + + K KE V G+K Y N+ D + EGV KVM++ G+
Sbjct 234 LCRRGEVEKSKKLIKEFSVKGIKPEAYTYNTIINAYVKQRDFSGVEGVLKVMKKDGV 290
> At4g04560
Length=590
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 18/66 (27%), Positives = 24/66 (36%), Gaps = 1/66 (1%)
Query 18 PMADDMHTHLRQVELMDMVTLQIRKGGCDRVLVMPNTVPPIVTCSQALEYRNELLKRDPN 77
P A T L D+ LQI + + P P + EY N +K N
Sbjct 50 PEAVQQKTRCEAASLKDLTALQILQTAVSDS-IFPRIAPASSALGKPWEYENLKMKESDN 108
Query 78 VNYLMT 83
+N MT
Sbjct 109 INTFMT 114
> At4g01080
Length=442
Score = 29.3 bits (64), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 24/44 (54%), Gaps = 3/44 (6%)
Query 51 MPNTVPPI---VTCSQALEYRNELLKRDPNVNYLMTLYLCREID 91
+P+ P+ VTC +++N LL P+VNYL + R+ D
Sbjct 99 IPDPTGPLYTNVTCRHIQDFQNCLLNGRPDVNYLFWRWKPRDCD 142
> At5g35820
Length=1342
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 24/87 (27%), Positives = 41/87 (47%), Gaps = 10/87 (11%)
Query 65 LEYRNELLKRDPNVNYLMTL--YLCREIDADDIAKRAKESHVVGVKLYPRGVTTNSEAGV 122
L Y +++L N + + L L RE + D+ K ++G++ +T N E G+
Sbjct 1019 LLYVDDMLIASQNKDQIQKLKESLNREFEMKDLGPARK---ILGME-----ITRNREQGI 1070
Query 123 EDLTQYEGVFKVMEELGMSLHIHAEKP 149
DL+Q E V V+ GM ++ P
Sbjct 1071 LDLSQSEYVAGVLRAFGMDQSKVSQTP 1097
> CE12384
Length=853
Score = 28.5 bits (62), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 22/36 (61%), Gaps = 2/36 (5%)
Query 38 LQIRKGGCDRVLVMPNTV--PPIVTCSQALEYRNEL 71
L +RK D+VLV+PNT+ PP C +E+ E+
Sbjct 287 LSVRKLILDKVLVLPNTISFPPSQVCDLLIEHDPEM 322
> CE20600
Length=1586
Score = 28.5 bits (62), Expect = 5.9, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 25/51 (49%), Gaps = 2/51 (3%)
Query 59 VTCSQALEYRNELLKRDPNVNYLMTLYLCREIDADDIAKRAKESHVVGVKL 109
VT Q +N LL D N NY +Y I++ I + ++ HVV V L
Sbjct 454 VTVLQCSRLKNALLLDDKNKNY--EVYCTVSIESRPIVQNEEQGHVVNVLL 502
> CE20599
Length=1584
Score = 28.5 bits (62), Expect = 6.2, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 25/51 (49%), Gaps = 2/51 (3%)
Query 59 VTCSQALEYRNELLKRDPNVNYLMTLYLCREIDADDIAKRAKESHVVGVKL 109
VT Q +N LL D N NY +Y I++ I + ++ HVV V L
Sbjct 454 VTVLQCSRLKNALLLDDKNKNY--EVYCTVSIESRPIVQNEEQGHVVNVLL 502
> YMR027w
Length=470
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 31/66 (46%), Gaps = 1/66 (1%)
Query 25 THLRQVELMDMVTLQIRKGGCDRVLVMPNTVPPIVTCSQALEYRNELLKRDPNVNYLMTL 84
T Q E++ + R+ DRV + P T I + L + NE LK+ P VN+
Sbjct 47 TKFEQGEVIKKELKEFRQEIIDRVPLRPFTEEEIKIANVPLSF-NEYLKKHPEVNWGAVE 105
Query 85 YLCREI 90
+L E+
Sbjct 106 WLFSEV 111
> At3g51320
Length=486
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 23/41 (56%), Gaps = 4/41 (9%)
Query 39 QIRKGGCDRVLVMPNTVPPIVTCSQALEYRNELL----KRD 75
Q K GCD+VL + N++ + TC AL+ +L KRD
Sbjct 120 QAIKHGCDQVLPVQNSLMHMYTCCGALDLAKKLFVEIPKRD 160
> 7298800
Length=568
Score = 28.1 bits (61), Expect = 6.8, Method: Composition-based stats.
Identities = 24/96 (25%), Positives = 41/96 (42%), Gaps = 12/96 (12%)
Query 67 YRNELLKRDPNVNYLMTLYLCREIDADDIAKRAKESHVVGVKLYPRGVTTNSEAGVEDLT 126
Y ELLK +P + LY ++ + A + G PRG+T ++ + DL
Sbjct 367 YMLELLKHEPAMVLPHELYQSMSVEERNTAADVLVKYHYG----PRGITKSNITQILDLF 422
Query 127 QYEGVFKVMEELGMSLHIHAEKP--------DAPTL 154
Y+ + + + +S HA+ P D+P L
Sbjct 423 SYKLFWHGIHRVVLSRLSHAQAPTYLYRFDFDSPKL 458
> CE23607
Length=1449
Score = 28.1 bits (61), Expect = 7.5, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 25/51 (49%), Gaps = 2/51 (3%)
Query 59 VTCSQALEYRNELLKRDPNVNYLMTLYLCREIDADDIAKRAKESHVVGVKL 109
VT Q +N LL D N NY +Y I++ I + ++ HVV V L
Sbjct 319 VTVLQCSRLKNALLLDDKNKNY--EVYCTVSIESRPIVQNEEQGHVVNVLL 367
Lambda K H
0.321 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2070320142
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40