bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2641_orf1
Length=149
Score E
Sequences producing significant alignments: (Bits) Value
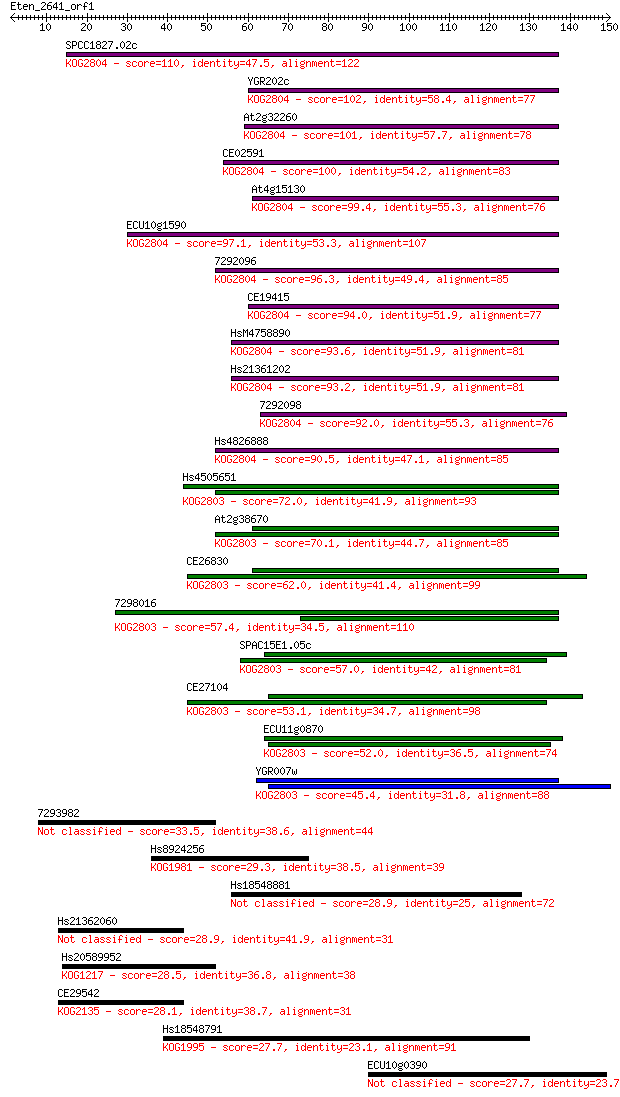
SPCC1827.02c 110 1e-24
YGR202c 102 2e-22
At2g32260 101 6e-22
CE02591 100 1e-21
At4g15130 99.4 3e-21
ECU10g1590 97.1 1e-20
7292096 96.3 2e-20
CE19415 94.0 1e-19
HsM4758890 93.6 1e-19
Hs21361202 93.2 1e-19
7292098 92.0 3e-19
Hs4826888 90.5 1e-18
Hs4505651 72.0 4e-13
At2g38670 70.1 2e-12
CE26830 62.0 4e-10
7298016 57.4 1e-08
SPAC15E1.05c 57.0 1e-08
CE27104 53.1 2e-07
ECU11g0870 52.0 4e-07
YGR007w 45.4 4e-05
7293982 33.5 0.18
Hs8924256 29.3 3.3
Hs18548881 28.9 3.4
Hs21362060 28.9 4.0
Hs20589952 28.5 4.8
CE29542 28.1 6.3
Hs18548791 27.7 8.6
ECU10g0390 27.7 9.1
> SPCC1827.02c
Length=354
Score = 110 bits (274), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 58/123 (47%), Positives = 78/123 (63%), Gaps = 7/123 (5%)
Query 15 PAAALEAGSSCTSPSMKKRKATEDPPSQPDPQGPAPGEFAEGEDPN-KPVRIYTDGVYDL 73
P+ +LE S SP E+P + P + + P +PVR+Y DGV+DL
Sbjct 51 PSKSLEEISQSVSP------VEEEPRDVRFKELSTPFSYPINDPPEGRPVRVYADGVFDL 104
Query 74 LHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETLRHIKWVDEVIA 133
H+GHMRQLEQAKK+F +V+L+ G+ +D+ THRLKG TV + +ERAE LRH KWVDEV+
Sbjct 105 FHIGHMRQLEQAKKVFPNVHLIVGLPNDQLTHRLKGLTVMNDKERAEALRHCKWVDEVLE 164
Query 134 PLP 136
P
Sbjct 165 NAP 167
> YGR202c
Length=424
Score = 102 bits (255), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 45/77 (58%), Positives = 59/77 (76%), Gaps = 0/77 (0%)
Query 60 NKPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERA 119
++P+RIY DGV+DL HLGHM+QLEQ KK F +V L+ GV SD+ TH+LKG TV + ++R
Sbjct 101 DRPIRIYADGVFDLFHLGHMKQLEQCKKAFPNVTLIVGVPSDKITHKLKGLTVLTDKQRC 160
Query 120 ETLRHIKWVDEVIAPLP 136
ETL H +WVDEV+ P
Sbjct 161 ETLTHCRWVDEVVPNAP 177
> At2g32260
Length=332
Score = 101 bits (251), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 45/79 (56%), Positives = 56/79 (70%), Gaps = 1/79 (1%)
Query 59 PNKPVRIYTDGVYDLLHLGHMRQLEQAKKMF-KHVYLMAGVASDEETHRLKGQTVQSLEE 117
++PVR+Y DG+YDL H GH R LEQAK F + YL+ G +DE TH+ KG+TV + EE
Sbjct 31 TDRPVRVYADGIYDLFHFGHARSLEQAKLAFPNNTYLLVGCCNDETTHKYKGRTVMTAEE 90
Query 118 RAETLRHIKWVDEVIAPLP 136
R E+LRH KWVDEVI P
Sbjct 91 RYESLRHCKWVDEVIPDAP 109
> CE02591
Length=347
Score = 100 bits (249), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 45/83 (54%), Positives = 57/83 (68%), Gaps = 0/83 (0%)
Query 54 AEGEDPNKPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQ 113
AE + +PVRIY DG+YDL H GH QL Q KKMF +VYL+ GV D +TH+ KG+TV
Sbjct 57 AEANEAGRPVRIYADGIYDLFHHGHANQLRQVKKMFPNVYLIVGVCGDRDTHKYKGRTVT 116
Query 114 SLEERAETLRHIKWVDEVIAPLP 136
S EER + +RH ++VDEV P
Sbjct 117 SEEERYDGVRHCRYVDEVYREAP 139
> At4g15130
Length=298
Score = 99.4 bits (246), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 42/76 (55%), Positives = 55/76 (72%), Gaps = 0/76 (0%)
Query 61 KPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAE 120
+PVR+Y DG++DL H GH R +EQAKK F + YL+ G +DE T++ KG+TV + ER E
Sbjct 19 RPVRVYADGIFDLFHFGHARAIEQAKKSFPNTYLLVGCCNDEITNKFKGKTVMTESERYE 78
Query 121 TLRHIKWVDEVIAPLP 136
+LRH KWVDEVI P
Sbjct 79 SLRHCKWVDEVIPDAP 94
> ECU10g1590
Length=278
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 57/122 (46%), Positives = 67/122 (54%), Gaps = 21/122 (17%)
Query 30 MKKRKATED----PPSQPDPQGPAPGEFAEGED-----------PNKPVRIYTDGVYDLL 74
MKK A+E+ PS PD E ED N+PVRIY DGVYDL
Sbjct 1 MKKDDASEEYRMMMPSYPDRNA------FEYEDMTTDVGMYRVPVNRPVRIYCDGVYDLF 54
Query 75 HLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETLRHIKWVDEVIAP 134
H GH R L+QAK +F +VYL+ GV D+ T RLKG V ERAE L H ++VDEVI
Sbjct 55 HYGHARSLKQAKNLFPNVYLLVGVTDDDITVRLKGNLVMDENERAEGLIHCRYVDEVITS 114
Query 135 LP 136
P
Sbjct 115 AP 116
> 7292096
Length=347
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 42/85 (49%), Positives = 58/85 (68%), Gaps = 0/85 (0%)
Query 52 EFAEGEDPNKPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQT 111
+ A + VR+Y DG+YDL H GH RQL QAK +F +VYL+ GV +DE THR+KG+T
Sbjct 16 QMARSGQTTRRVRVYADGIYDLFHQGHARQLMQAKNIFPNVYLIVGVCNDELTHRMKGRT 75
Query 112 VQSLEERAETLRHIKWVDEVIAPLP 136
V + ER E +RH ++VDE++ P
Sbjct 76 VMNGFERYEGVRHCRYVDEIVQNAP 100
> CE19415
Length=183
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 40/77 (51%), Positives = 55/77 (71%), Gaps = 0/77 (0%)
Query 60 NKPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERA 119
++P+R+Y DG+YD+ H GH +QL Q K+MF VYL+ GV SDE T + KG TVQS ER
Sbjct 39 SRPIRVYADGIYDMFHYGHAKQLLQIKQMFPMVYLIVGVCSDENTLKFKGPTVQSENERY 98
Query 120 ETLRHIKWVDEVIAPLP 136
E++R ++VDEV+ P
Sbjct 99 ESVRQCRYVDEVLKDAP 115
> HsM4758890
Length=330
Score = 93.6 bits (231), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 42/81 (51%), Positives = 55/81 (67%), Gaps = 0/81 (0%)
Query 56 GEDPNKPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSL 115
G ++PVR+Y DG++DL H GH R L QAK +F + YL+ GV SD+ TH+ KG TV +
Sbjct 70 GTPADRPVRVYADGIFDLFHSGHARALMQAKTLFPNSYLLVGVCSDDLTHKFKGFTVMNE 129
Query 116 EERAETLRHIKWVDEVIAPLP 136
ER E LRH ++VDEVI P
Sbjct 130 AERYEALRHCRYVDEVIRDAP 150
> Hs21361202
Length=369
Score = 93.2 bits (230), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 42/81 (51%), Positives = 55/81 (67%), Gaps = 0/81 (0%)
Query 56 GEDPNKPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSL 115
G ++PVR+Y DG++DL H GH R L QAK +F + YL+ GV SD+ TH+ KG TV +
Sbjct 70 GTPADRPVRVYADGIFDLFHSGHARALMQAKTLFPNSYLLVGVCSDDLTHKFKGFTVMNE 129
Query 116 EERAETLRHIKWVDEVIAPLP 136
ER E LRH ++VDEVI P
Sbjct 130 AERYEALRHCRYVDEVIRDAP 150
> 7292098
Length=381
Score = 92.0 bits (227), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 42/78 (53%), Positives = 57/78 (73%), Gaps = 2/78 (2%)
Query 63 VRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETL 122
VR+Y DG+YDL H GH RQL QAK +F +VYL+ GV +DE T R+KG+TV + ER E +
Sbjct 79 VRVYADGIYDLFHQGHARQLMQAKNVFPNVYLIVGVCNDELTLRMKGRTVMNGFERYEAV 138
Query 123 RHIKWVDEVI--APLPLD 138
RH ++VDE++ AP L+
Sbjct 139 RHCRYVDEIVPNAPWTLN 156
> Hs4826888
Length=367
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 40/85 (47%), Positives = 55/85 (64%), Gaps = 0/85 (0%)
Query 52 EFAEGEDPNKPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQT 111
E + G +PVR+Y DG++DL H GH R L QAK +F + YL+ GV SDE TH KG T
Sbjct 66 EASRGTPCERPVRVYADGIFDLFHSGHARALMQAKNLFPNTYLIVGVCSDELTHNFKGFT 125
Query 112 VQSLEERAETLRHIKWVDEVIAPLP 136
V + ER + ++H ++VDEV+ P
Sbjct 126 VMNENERYDAVQHCRYVDEVVRNAP 150
> Hs4505651
Length=389
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 39/93 (41%), Positives = 55/93 (59%), Gaps = 2/93 (2%)
Query 44 DPQGPAPGEFAEGEDPNKPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEE 103
+ +G A G G + VR++ DG YD++H GH QL QA+ M YL+ GV +DEE
Sbjct 4 NGRGAAGGAEQPGPGGRRAVRVWCDGCYDMVHYGHSNQLRQARAMGD--YLIVGVHTDEE 61
Query 104 THRLKGQTVQSLEERAETLRHIKWVDEVIAPLP 136
+ KG V + EER + ++ IKWVDEV+ P
Sbjct 62 IAKHKGPPVFTQEERYKMVQAIKWVDEVVPAAP 94
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 51/88 (57%), Gaps = 3/88 (3%)
Query 52 EFAEGEDPN-KPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQ 110
+FA G++P IY G +DL H+GH+ LE+ ++ + Y++AG+ D+E + KG+
Sbjct 202 QFASGKEPQPGETVIYVAGAFDLFHIGHVDFLEKVHRLAERPYIIAGLHFDQEVNHYKGK 261
Query 111 T--VQSLEERAETLRHIKWVDEVIAPLP 136
+ +L ER ++ ++V EV+ P
Sbjct 262 NYPIMNLHERTLSVLACRYVSEVVIGAP 289
> At2g38670
Length=421
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 35/76 (46%), Positives = 46/76 (60%), Gaps = 2/76 (2%)
Query 61 KPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAE 120
KPVR+Y DG +D++H GH L QA+ + L+ GV SDEE KG V L ER
Sbjct 53 KPVRVYMDGCFDMMHYGHCNALRQARALGDQ--LVVGVVSDEEIIANKGPPVTPLHERMT 110
Query 121 TLRHIKWVDEVIAPLP 136
++ +KWVDEVI+ P
Sbjct 111 MVKAVKWVDEVISDAP 126
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 51/88 (57%), Gaps = 5/88 (5%)
Query 52 EFAEGEDPNKPVRI-YTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKG- 109
+F+ G+ P RI Y DG +DL H GH+ L +A+++ +L+ G+ +D+ +G
Sbjct 243 QFSNGKGPGPDARIIYIDGAFDLFHAGHVEILRRARELGD--FLLVGIHNDQTVSAKRGA 300
Query 110 -QTVQSLEERAETLRHIKWVDEVIAPLP 136
+ + +L ER+ ++ ++VDEVI P
Sbjct 301 HRPIMNLHERSLSVLACRYVDEVIIGAP 328
> CE26830
Length=370
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 33/76 (43%), Positives = 45/76 (59%), Gaps = 2/76 (2%)
Query 61 KPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAE 120
K R++ DG YD++H GH QL QAK+ + L+ GV +DEE KG V + +ER
Sbjct 11 KGNRVWADGCYDMVHFGHANQLRQAKQFGQK--LIVGVHNDEEIRLHKGPPVFNEQERYR 68
Query 121 TLRHIKWVDEVIAPLP 136
+ IKWVDEV+ P
Sbjct 69 MVAGIKWVDEVVENAP 84
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 36/103 (34%), Positives = 55/103 (53%), Gaps = 7/103 (6%)
Query 45 PQGPAPGEFAEGEDPNKPVR--IYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDE 102
P EFAEG P KP +Y G +DL H+GH+ LE+AK+ YL+ G+ SD+
Sbjct 185 PTTTTILEFAEGRPP-KPTDKVVYVTGSFDLFHIGHLAFLEKAKEFGD--YLIVGILSDQ 241
Query 103 ETHRLKGQT--VQSLEERAETLRHIKWVDEVIAPLPLDHHSRV 143
++ KG + S+ ER ++ K V+EV+ P + S +
Sbjct 242 TVNQYKGSNHPIMSIHERVLSVLAYKPVNEVVFGAPYEITSDI 284
> 7298016
Length=363
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 38/116 (32%), Positives = 62/116 (53%), Gaps = 8/116 (6%)
Query 27 SPSMKKRKATEDP---PSQPDPQGPAPGEFAEGEDPNKPVRI-YTDGVYDLLHLGHMRQL 82
S +M + A + P SQ P +F++G+ PN +I Y G +DL H+GH+ L
Sbjct 155 SSNMGQDSAAKSPWTGCSQFLPTTQKIIQFSDGKSPNPGDKIVYVAGAFDLFHVGHLDFL 214
Query 83 EQAKKMFKHVYLMAGVASDEETHRLKGQT--VQSLEERAETLRHIKWVDEVIAPLP 136
E+AKK+ YL+ G+ +D + KG + +L ER ++ K+V+EV+ P
Sbjct 215 EKAKKLGD--YLIVGLHTDPVVNSYKGSNYPIMNLHERVLSVLACKFVNEVVIGAP 268
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 38/64 (59%), Gaps = 2/64 (3%)
Query 73 LLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETLRHIKWVDEVI 132
++H GH L QAK + V + G+ +DEE + KG V + EER + ++ IKWVDEV+
Sbjct 1 MVHFGHANSLRQAKALGDKV--IVGIHTDEEITKHKGPPVFTEEERVKMVKGIKWVDEVV 58
Query 133 APLP 136
P
Sbjct 59 LGAP 62
> SPAC15E1.05c
Length=365
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 32/77 (41%), Positives = 43/77 (55%), Gaps = 4/77 (5%)
Query 64 RIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETLR 123
R++ DG D H GH + QAK++ + L+ G+ SDEE KG V +LEER +
Sbjct 10 RLWLDGCMDFFHYGHSNAILQAKQLGET--LVIGIHSDEEITLNKGPPVMTLEERCLSAN 67
Query 124 HIKWVDEVI--APLPLD 138
KWVDEV+ AP D
Sbjct 68 TCKWVDEVVPSAPYVFD 84
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 40/76 (52%), Gaps = 4/76 (5%)
Query 58 DPNKPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEE 117
+P K + IY DG +DL H+ LE +MF + +MAG+ +DE+ + + +L E
Sbjct 199 NPEKNI-IYIDGDWDLFTEKHISALELCTRMFPGIPIMAGIFADEKCFE---KPMLNLLE 254
Query 118 RAETLRHIKWVDEVIA 133
R L K++ ++
Sbjct 255 RILNLLQCKYISSILV 270
> CE27104
Length=302
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 43/80 (53%), Gaps = 4/80 (5%)
Query 65 IYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQT--VQSLEERAETL 122
+Y G +DL H GH+ LE AK + YL+ G+ D++ + KG V +L ER +
Sbjct 151 VYVSGAFDLFHAGHLSFLEAAKDLGD--YLIVGIVGDDDVNEEKGTIFPVMNLLERTLNI 208
Query 123 RHIKWVDEVIAPLPLDHHSR 142
+K VDEV +P +S+
Sbjct 209 SSLKIVDEVFVGVPAVTNSK 228
Score = 51.2 bits (121), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 32/89 (35%), Positives = 46/89 (51%), Gaps = 4/89 (4%)
Query 45 PQGPAPGEFAEGEDPNKPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEET 104
P+ G +GE K R+YTDG +D +H + R L AK+ K L+ G+ SD+E
Sbjct 2 PKAIVQGLTPDGE--KKKARVYTDGCFDFVHFANARLLWPAKQYGKK--LIVGIHSDDEL 57
Query 105 HRLKGQTVQSLEERAETLRHIKWVDEVIA 133
+ + EER + I+WVDE IA
Sbjct 58 DNNGILPIFTDEERYRLISAIRWVDEEIA 86
> ECU11g0870
Length=322
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 41/74 (55%), Gaps = 2/74 (2%)
Query 64 RIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETLR 123
+++ DG +D+ H GH L Q+K + YL+AGV S ++ KG V EER E +
Sbjct 8 KVWADGCFDMFHYGHANALRQSKALGD--YLIAGVHSSLSINQEKGLPVMEDEERYEVVE 65
Query 124 HIKWVDEVIAPLPL 137
++VDEV+ P
Sbjct 66 GCRYVDEVVRDAPF 79
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 38/71 (53%), Gaps = 3/71 (4%)
Query 65 IYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRL-KGQTVQSLEERAETLR 123
++ DG +DL H GH+ L A+ M YL+ G+ DE T + V S +ER TL
Sbjct 180 VFMDGNFDLFHAGHVASLRIARGMGD--YLIVGIHDDETTKEYTRSYPVLSTKERMLTLM 237
Query 124 HIKWVDEVIAP 134
++VDE++
Sbjct 238 ACRYVDEIVVS 248
> YGR007w
Length=323
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 39/77 (50%), Gaps = 2/77 (2%)
Query 62 PVRIYTDGVYDLLHLGHMRQLEQAKKMF--KHVYLMAGVASDEETHRLKGQTVQSLEERA 119
P +++ DG +D H GH + QA++ ++ L GV +DE+ KG V + ER
Sbjct 7 PDKVWIDGCFDFTHHGHAGAILQARRTVSKENGKLFCGVHTDEDIQHNKGTPVMNSSERY 66
Query 120 ETLRHIKWVDEVIAPLP 136
E R +W EV+ P
Sbjct 67 EHTRSNRWCSEVVEAAP 83
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 25/87 (28%), Positives = 43/87 (49%), Gaps = 8/87 (9%)
Query 65 IYTDGVYDLLHLGHMRQLEQAKKMFKH--VYLMAGVASDEETHRLKGQTVQSLEERAETL 122
+Y DG +DL H+G + QL + KM H L+ G+ + + + T+ +++ER ++
Sbjct 200 VYVDGDFDLFHMGDIDQLRKL-KMDLHPDKKLIVGITTSDYS-----STIMTMKERVLSV 253
Query 123 RHIKWVDEVIAPLPLDHHSRVRREAPH 149
K+VD VI S+ E H
Sbjct 254 LSCKYVDAVIIDADATSMSQYNCEKYH 280
> 7293982
Length=1322
Score = 33.5 bits (75), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 26/44 (59%), Gaps = 0/44 (0%)
Query 8 PSRGLWLPAAALEAGSSCTSPSMKKRKATEDPPSQPDPQGPAPG 51
PS G P A+ +S TSP +++R++T PP+ P+ G A G
Sbjct 1199 PSTGAGSPPASSPKATSSTSPKIQRRRSTSKPPTTPNGLGSATG 1242
> Hs8924256
Length=441
Score = 29.3 bits (64), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 20/45 (44%), Gaps = 6/45 (13%)
Query 36 TEDPPSQPDPQ------GPAPGEFAEGEDPNKPVRIYTDGVYDLL 74
T PP+ PD GP+P E A +P P + G +LL
Sbjct 194 TMSPPTCPDTSDSSSVAGPSPNEAANNPEPLSPTMVLCQGFLNLL 238
> Hs18548881
Length=549
Score = 28.9 bits (63), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 18/72 (25%), Positives = 30/72 (41%), Gaps = 2/72 (2%)
Query 56 GEDPNKPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSL 115
G+ P+ +Y + L +L H +L + ++ + S+ T R K +Q L
Sbjct 392 GQSAEPPISVYF--LPALSYLPHQLRLTCMNSSLNSMSCLSQLMSESATCRAKSSLIQGL 449
Query 116 EERAETLRHIKW 127
EE E R W
Sbjct 450 EENEERRRLFAW 461
> Hs21362060
Length=147
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 16/31 (51%), Gaps = 0/31 (0%)
Query 13 WLPAAALEAGSSCTSPSMKKRKATEDPPSQP 43
W P A AG SC +P+ + PPSQP
Sbjct 104 WEPGGAPGAGPSCAAPAAAEVGGPRGPPSQP 134
> Hs20589952
Length=737
Score = 28.5 bits (62), Expect = 4.8, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 18/38 (47%), Gaps = 1/38 (2%)
Query 14 LPAAALEAGSSCTSPSMKKRKATEDPPSQPDPQGPAPG 51
+PAA L A C + + P +PDPQ PAP
Sbjct 37 VPAAPLSAPGPCAAQPCRNGGVCTSRP-EPDPQHPAPA 73
> CE29542
Length=719
Score = 28.1 bits (61), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 13 WLPAAALEAGSSCTSPSMKKRKATEDPPSQP 43
++PA+ +S T+P +KK KA + PS+P
Sbjct 78 YMPASTAPTSASATAPDLKKEKAPAEKPSKP 108
> Hs18548791
Length=361
Score = 27.7 bits (60), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 21/92 (22%), Positives = 37/92 (40%), Gaps = 11/92 (11%)
Query 39 PPSQPDPQGPAPGEFAEGEDPNKPVRIYTDGVYDLL-HLGHMRQLEQAKKMFKHVYLMAG 97
PP PD + +G + N + D + D G ++ ++ ++ H YL
Sbjct 76 PPVDPDEDSDNSAIYVQGLNDN----VTLDDLVDFFKQCGVVKMNKRTEQPMIHTYL--- 128
Query 98 VASDEETHRLKGQTVQSLEERAETLRHIKWVD 129
D+ET + KG S E+ ++W D
Sbjct 129 ---DKETRKPKGDATVSCEDSPTAKAAVEWFD 157
> ECU10g0390
Length=514
Score = 27.7 bits (60), Expect = 9.1, Method: Composition-based stats.
Identities = 14/59 (23%), Positives = 28/59 (47%), Gaps = 0/59 (0%)
Query 90 KHVYLMAGVASDEETHRLKGQTVQSLEERAETLRHIKWVDEVIAPLPLDHHSRVRREAP 148
K +Y M + T R+ G + + +A ++ + V +I +P++H +RR P
Sbjct 412 KDLYRMIARYMENHTTRIVGLITEMIGRKASDVQILMEVVRIIEEMPMEHLRIIRRIKP 470
Lambda K H
0.313 0.132 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1858150626
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40