bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2732_orf1
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
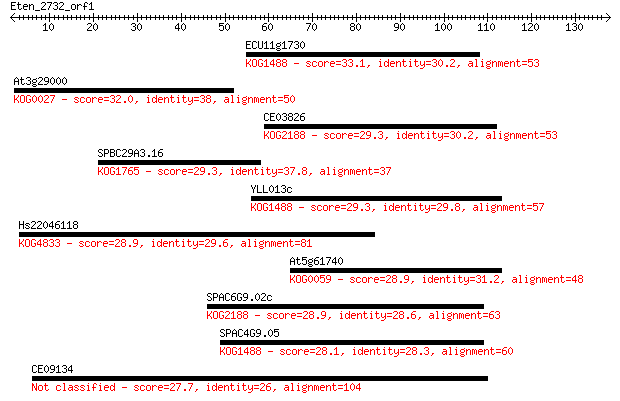
ECU11g1730 33.1 0.15
At3g29000 32.0 0.35
CE03826 29.3 2.5
SPBC29A3.16 29.3 2.5
YLL013c 29.3 2.5
Hs22046118 28.9 2.9
At5g61740 28.9 2.9
SPAC6G9.02c 28.9 3.1
SPAC4G9.05 28.1 5.1
CE09134 27.7 7.5
> ECU11g1730
Length=530
Score = 33.1 bits (74), Expect = 0.15, Method: Composition-based stats.
Identities = 16/53 (30%), Positives = 32/53 (60%), Gaps = 1/53 (1%)
Query 55 ALSESACGSRTLEVFFSFFSSQTFQKFFSSFLLKRAPQLAAGRFGNFLLQKVL 107
++S+ GSR ++ S FF++ ++ AP+L+A FGN+++QK++
Sbjct 224 SISKDQEGSRCIQRKMDSISRAEISWFFNN-IVDAAPELSANLFGNYVIQKII 275
> At3g29000
Length=194
Score = 32.0 bits (71), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 26/51 (50%), Gaps = 1/51 (1%)
Query 2 LFTRFNFFFVSFC-FIFFNSLFAAAAEPLQQLVEALFSSPKEKDSDEELVK 51
LF FNFF +SFC ++ +F + PL Q + +F KD E L K
Sbjct 12 LFALFNFFLISFCRWVSSTRIFLSRFVPLLQHHQRVFDKKNNKDQQETLTK 62
> CE03826
Length=630
Score = 29.3 bits (64), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 23/53 (43%), Gaps = 0/53 (0%)
Query 59 SACGSRTLEVFFSFFSSQTFQKFFSSFLLKRAPQLAAGRFGNFLLQKVLQSPS 111
S+ GSR + S + F K +L RF NF LQK++ S +
Sbjct 353 SSNGSRVWDKLMETCSEDARSLLWIEFCSKNVDELTDNRFSNFPLQKMINSST 405
> SPBC29A3.16
Length=166
Score = 29.3 bits (64), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 24/37 (64%), Gaps = 0/37 (0%)
Query 21 LFAAAAEPLQQLVEALFSSPKEKDSDEELVKFAEALS 57
LF+ + + +QQLV + S PKE+ SD L++ E ++
Sbjct 40 LFSLSRDNVQQLVNKMISLPKERTSDGVLLQLPETVT 76
> YLL013c
Length=879
Score = 29.3 bits (64), Expect = 2.5, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 28/57 (49%), Gaps = 1/57 (1%)
Query 56 LSESACGSRTLEVFFSFFSSQTFQKFFSSFLLKRAPQLAAGRFGNFLLQKVLQSPSF 112
LS + G R ++ F SS+ + + L P L ++GN+++Q VLQ F
Sbjct 690 LSTHSYGCRVIQRLLEFGSSEDQESILNE-LKDFIPYLIQDQYGNYVIQYVLQQDQF 745
> Hs22046118
Length=1767
Score = 28.9 bits (63), Expect = 2.9, Method: Composition-based stats.
Identities = 24/82 (29%), Positives = 33/82 (40%), Gaps = 1/82 (1%)
Query 3 FTRFNFFF-VSFCFIFFNSLFAAAAEPLQQLVEALFSSPKEKDSDEELVKFAEALSESAC 61
+NF F +SF F+S A + L V + E D +E + A LS A
Sbjct 1601 LAEYNFLFALSFTTPTFDSEVAPSFGTLLATVNVALNMLGELDKKKEPLTQAVGLSTQAE 1660
Query 62 GSRTLEVFFSFFSSQTFQKFFS 83
G+RTL+ F F S
Sbjct 1661 GTRTLKSLLMFTMENCFYLLIS 1682
> At5g61740
Length=848
Score = 28.9 bits (63), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 28/48 (58%), Gaps = 0/48 (0%)
Query 65 TLEVFFSFFSSQTFQKFFSSFLLKRAPQLAAGRFGNFLLQKVLQSPSF 112
L++ F+F +S F K ++ ++ A+G G FL ++L+SP+F
Sbjct 353 NLQISFAFLASSIFSKVKTATVVAYTLVFASGLLGMFLFGELLESPTF 400
> SPAC6G9.02c
Length=655
Score = 28.9 bits (63), Expect = 3.1, Method: Composition-based stats.
Identities = 18/63 (28%), Positives = 31/63 (49%), Gaps = 1/63 (1%)
Query 46 DEELVKFAEALSESACGSRTLEVFFSFFSSQTFQKFFSSFLLKRAPQLAAGRFGNFLLQK 105
D + + F E L + GSR LEV S+ +F++ F R +L NF++Q+
Sbjct 312 DLKELPFMETLLKDETGSRILEVIVENMSASHLLRFYAVF-EGRFYRLCVHPIANFIMQR 370
Query 106 VLQ 108
++
Sbjct 371 YIR 373
> SPAC4G9.05
Length=581
Score = 28.1 bits (61), Expect = 5.1, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 32/60 (53%), Gaps = 1/60 (1%)
Query 49 LVKFAEALSESACGSRTLEVFFSFFSSQTFQKFFSSFLLKRAPQLAAGRFGNFLLQKVLQ 108
++ F+ L ++ G+ ++ F + + Q F S FL K +L+ FG+ +LQK L+
Sbjct 292 ILPFSVTLMKNKFGNFLIQKCFEYSTEAQLQSF-SYFLKKHVKELSIDAFGSHVLQKSLE 350
> CE09134
Length=624
Score = 27.7 bits (60), Expect = 7.5, Method: Composition-based stats.
Identities = 27/116 (23%), Positives = 49/116 (42%), Gaps = 12/116 (10%)
Query 6 FNFFFVSFCFIFFNSLFAAAAEPLQQLVEALFSSPK------EKDSDE-ELVKFAEALSE 58
+ FF++S FI FN++FA + L ++ P +D +E E + +E + E
Sbjct 179 YAFFYISQDFIAFNNVFARIWQFLVGMIVYTIGLPNPQYQVINQDIEECENLIDSEEVPE 238
Query 59 SACGSRTLEVFFSFFSSQTFQKFFSSFLLKRAPQLAAGRF-----GNFLLQKVLQS 109
+ C + + F+ F +++ + G F GNFL+ L S
Sbjct 239 TPCNMYSYFLLLGLFTVAAFPLKLHPLIVRPLVTVGTGSFILISEGNFLVSNKLIS 294
Lambda K H
0.329 0.140 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1461889840
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40