bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2735_orf1
Length=149
Score E
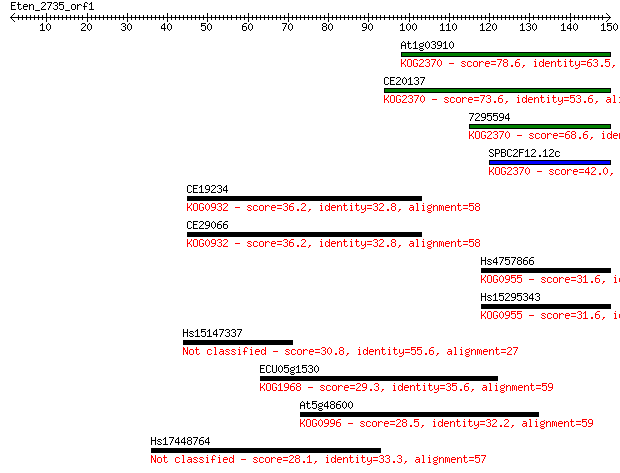
Sequences producing significant alignments: (Bits) Value
At1g03910 78.6 4e-15
CE20137 73.6 1e-13
7295594 68.6 4e-12
SPBC2F12.12c 42.0 4e-04
CE19234 36.2 0.022
CE29066 36.2 0.023
Hs4757866 31.6 0.54
Hs15295343 31.6 0.54
Hs15147337 30.8 1.1
ECU05g1530 29.3 3.0
At5g48600 28.5 5.1
Hs17448764 28.1 6.9
> At1g03910
Length=672
Score = 78.6 bits (192), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 33/53 (62%), Positives = 41/53 (77%), Gaps = 1/53 (1%)
Query 98 VMADSSEMKL-KQHYGWEEKYRPRKPRFFNRVRTCYFWNKYNQTHYDHDNPPP 149
+ ++E+ L + Y W +KYRPRKP++FNRV T Y WNKYNQTHYDHDNPPP
Sbjct 532 IFGSNAEVNLDSEVYWWHDKYRPRKPKYFNRVHTGYEWNKYNQTHYDHDNPPP 584
> CE20137
Length=667
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 30/56 (53%), Positives = 39/56 (69%), Gaps = 1/56 (1%)
Query 94 PDEEVMADSSEMKLKQHYGWEEKYRPRKPRFFNRVRTCYFWNKYNQTHYDHDNPPP 149
DE + ++ ++H W +KYRPRKP + NRV+T + WNKYNQTHYD DNPPP
Sbjct 526 ADESIFGAEEQLAAQRHL-WSDKYRPRKPTYLNRVQTGFDWNKYNQTHYDQDNPPP 580
> 7295594
Length=720
Score = 68.6 bits (166), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 28/35 (80%), Positives = 31/35 (88%), Gaps = 0/35 (0%)
Query 115 EKYRPRKPRFFNRVRTCYFWNKYNQTHYDHDNPPP 149
+KYRPRKPR+FNRV T + WNKYNQTHYD DNPPP
Sbjct 598 DKYRPRKPRYFNRVHTGFEWNKYNQTHYDMDNPPP 632
> SPBC2F12.12c
Length=517
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 120 RKPRFFNRVRTCYFWNKYNQTHYDHDNPPP 149
+KP +FNRV + WN YNQ H++ +PPP
Sbjct 389 KKPHYFNRVLLGFEWNSYNQAHFNEAHPPP 418
> CE19234
Length=816
Score = 36.2 bits (82), Expect = 0.022, Method: Composition-based stats.
Identities = 19/58 (32%), Positives = 32/58 (55%), Gaps = 5/58 (8%)
Query 45 ERERRLIQLGIDKDKNEKEEEENDDEILDPAAKIVTYEDFVRKEQKNASPDEEVMADS 102
E E + + D+ +N+K+ ENDDE++ +V E F+ SP+EE++A S
Sbjct 142 ESEEETVMMTTDEKENQKKPNENDDEVM-----VVDEEQFIVVSNDMKSPNEEIVAKS 194
> CE29066
Length=818
Score = 36.2 bits (82), Expect = 0.023, Method: Composition-based stats.
Identities = 19/58 (32%), Positives = 32/58 (55%), Gaps = 5/58 (8%)
Query 45 ERERRLIQLGIDKDKNEKEEEENDDEILDPAAKIVTYEDFVRKEQKNASPDEEVMADS 102
E E + + D+ +N+K+ ENDDE++ +V E F+ SP+EE++A S
Sbjct 144 ESEEETVMMTTDEKENQKKPNENDDEVM-----VVDEEQFIVVSNDMKSPNEEIVAKS 196
> Hs4757866
Length=1214
Score = 31.6 bits (70), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 20/38 (52%), Gaps = 6/38 (15%)
Query 118 RPRKPRFFNRVRTC------YFWNKYNQTHYDHDNPPP 149
R KP + V TC Y +Y+ HYDHDNPPP
Sbjct 15 RATKPPYECPVETCRKVYKSYSGIEYHLYHYDHDNPPP 52
> Hs15295343
Length=1214
Score = 31.6 bits (70), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 20/38 (52%), Gaps = 6/38 (15%)
Query 118 RPRKPRFFNRVRTC------YFWNKYNQTHYDHDNPPP 149
R KP + V TC Y +Y+ HYDHDNPPP
Sbjct 15 RATKPPYECPVETCRKVYKSYSGIEYHLYHYDHDNPPP 52
> Hs15147337
Length=2799
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 15/27 (55%), Positives = 19/27 (70%), Gaps = 3/27 (11%)
Query 44 RERERRLIQLGIDKDKNEKEEEENDDE 70
R+R R L++LGID NE E ENDD+
Sbjct 1974 RKRTRELLELGID---NEDSEHENDDD 1997
> ECU05g1530
Length=568
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 21/59 (35%), Positives = 27/59 (45%), Gaps = 8/59 (13%)
Query 63 EEEENDDEILDPAAKIVTYEDFVRKEQKNASPDEEVMADSSEMKLKQHYGWEEKYRPRK 121
+EEE D I D + + + +K +K AS DE A S W EKYRP K
Sbjct 71 DEEEFDSLIKDTSKALDGTAEVEKKTEKAASRDECGSASSGV--------WSEKYRPSK 121
> At5g48600
Length=1241
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 29/59 (49%), Gaps = 1/59 (1%)
Query 73 DPAAKIVTYEDFVRKEQKNASPDEEVMADSSEMKLKQHYGWEEKYRPRKPRFFNRVRTC 131
D AKI D ++ + N+ DE V D S +LK+ EK++ R+ N +R C
Sbjct 271 DTVAKITEQRDSLQNLE-NSLKDERVKMDESNEELKKFESVHEKHKKRQEVLDNELRAC 328
> Hs17448764
Length=364
Score = 28.1 bits (61), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 35/58 (60%), Gaps = 1/58 (1%)
Query 36 NERRIAKHRERERRLIQLGIDKDKNEKE-EEENDDEILDPAAKIVTYEDFVRKEQKNA 92
N+ R+ +E+ R L + KD+N E +EE+DD+I DP + + E++ +E ++A
Sbjct 128 NKFRVVIFQEKFRCPTSLSVVKDRNLTENQEEDDDDIFDPPVDLSSDEEYYVEESRSA 185
Lambda K H
0.315 0.135 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1858150626
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40