bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2737_orf1
Length=348
Score E
Sequences producing significant alignments: (Bits) Value
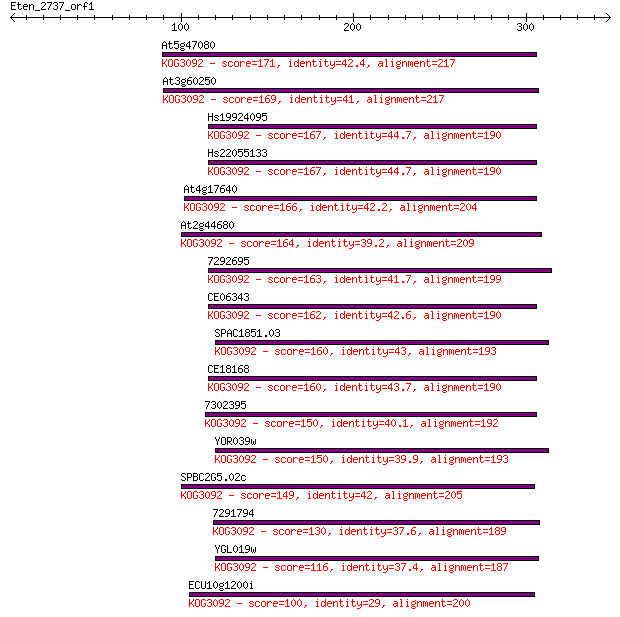
At5g47080 171 2e-42
At3g60250 169 7e-42
Hs19924095 167 3e-41
Hs22055133 167 3e-41
At4g17640 166 6e-41
At2g44680 164 2e-40
7292695 163 6e-40
CE06343 162 1e-39
SPAC1851.03 160 3e-39
CE18168 160 3e-39
7302395 150 4e-36
YOR039w 150 5e-36
SPBC2G5.02c 149 1e-35
7291794 130 3e-30
YGL019w 116 8e-26
ECU10g1200i 100 3e-21
> At5g47080
Length=287
Score = 171 bits (432), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 92/225 (40%), Positives = 137/225 (60%), Gaps = 13/225 (5%)
Query 89 STVASMFLKGSEEEEEEEEEEEEEE----EEEEEGWISWFCSLEGHDCFAEVDEEYIRDT 144
S AS+ + ++E + + EE + + E+ WISWFC+L G++ F EVD++YI+D
Sbjct 66 SRSASLSKNNTVSDDESDTDSEESDVSGSDGEDTSWISWFCNLRGNEFFCEVDDDYIQDD 125
Query 145 FNLFGLKSAVNNFEGAMDMILGDAPDEEEVADQSFMDIYRDAVD-LYGLIHARYIISPRG 203
FNL GL S V +E A+D+IL + E+ + ++ A + LYGLIHARYI++ +G
Sbjct 126 FNLCGLSSLVPYYEYALDLILDVESSQGEMFTEEQNELIESAAEMLYGLIHARYILTSKG 185
Query 204 LPQMRDRCIDEIFGHCPRVLCDRQVLLPVGISPDLRLHKLQLFCPLCQEVYDMVNAHERT 263
L M D+ + FG CPRV C Q LPVG S R ++++CP C+++Y + R+
Sbjct 186 LAAMLDKYKNYDFGRCPRVYCCGQPCLPVGQSDLPRSSTVKIYCPKCEDIY-----YPRS 240
Query 264 K---DIDGAYFGPSFPHIFLQTYSSWVPSVAPIPYQPRLFGFRIH 305
K +IDGAYFG +FPH+FL TY P+ A Y R+FGF++H
Sbjct 241 KYQGNIDGAYFGTTFPHLFLMTYGHLKPAKATQNYVQRVFGFKLH 285
> At3g60250
Length=276
Score = 169 bits (428), Expect = 7e-42, Method: Compositional matrix adjust.
Identities = 89/221 (40%), Positives = 129/221 (58%), Gaps = 9/221 (4%)
Query 90 TVASMFLKGSEEEEEEEEEEEEEEEEEEEGWISWFCSLEGHDCFAEVDEEYIRDTFNLFG 149
T L E E + E + E ++ WISWFC+L G+D F EVDE+YI+D FNL G
Sbjct 60 TSTKTQLPDVESETDSEGSDVSGSEGDDTSWISWFCNLRGNDFFCEVDEDYIQDDFNLCG 119
Query 150 LKSAVNNFEGAMDMILG-DAPDEEEVADQSFMDIYRDAVDLYGLIHARYIISPRGLPQMR 208
L V ++ A+D+IL DA + E D+ + A LYGLIH RYI++ +G+ M
Sbjct 120 LSGQVPYYDYALDLILDVDASNSEMFTDEQHEMVESAAEMLYGLIHVRYILTTKGMAAMT 179
Query 209 DRCIDEIFGHCPRVLCDRQVLLPVGISPDLRLHKLQLFCPLCQEVYDMVNAHERTK---D 265
++ + FG CPRV C Q LPVG S R ++++CP C+++ ++ R+K +
Sbjct 180 EKYKNCDFGRCPRVFCCGQSCLPVGQSDIPRSSTVKIYCPKCEDI-----SYPRSKFQGN 234
Query 266 IDGAYFGPSFPHIFLQTYSSWVPSVAPIPYQPRLFGFRIHN 306
IDGAYFG +FPH+FL TY + P Y P++FGF++H
Sbjct 235 IDGAYFGTTFPHLFLMTYGNLKPQKPTQSYVPKIFGFKVHK 275
> Hs19924095
Length=215
Score = 167 bits (423), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 85/191 (44%), Positives = 118/191 (61%), Gaps = 3/191 (1%)
Query 116 EEEGWISWFCSLEGHDCFAEVDEEYIRDTFNLFGLKSAVNNFEGAMDMILGDAPDEEEVA 175
EE WISWFC L G++ F EVDE+YI+D FNL GL V ++ A+DMIL PDEE
Sbjct 5 EEVSWISWFCGLRGNEFFCEVDEDYIQDKFNLTGLNEQVPHYRQALDMILDLEPDEELED 64
Query 176 DQSFMDIYRDAVD-LYGLIHARYIISPRGLPQMRDRCIDEIFGHCPRVLCDRQVLLPVGI 234
+ + D+ A + LYGLIHARYI++ RG+ QM ++ FG+CPRV C+ Q +LP+G+
Sbjct 65 NPNQSDLIEQAAEMLYGLIHARYILTNRGIAQMLEKYQQGDFGYCPRVYCENQPMLPIGL 124
Query 235 SPDLRLHKLQLFCPLCQEVYDMVNAHERTKDIDGAYFGPSFPHIFLQTYSSWVPSVAPIP 294
S ++L+CP C +VY ++ R DGAYFG FPH+ + + P
Sbjct 125 SDIPGEAMVKLYCPKCMDVYTPKSS--RHHHTDGAYFGTGFPHMLFMVHPEYRPKRPANQ 182
Query 295 YQPRLFGFRIH 305
+ PRL+GF+IH
Sbjct 183 FVPRLYGFKIH 193
> Hs22055133
Length=215
Score = 167 bits (423), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 85/191 (44%), Positives = 118/191 (61%), Gaps = 3/191 (1%)
Query 116 EEEGWISWFCSLEGHDCFAEVDEEYIRDTFNLFGLKSAVNNFEGAMDMILGDAPDEEEVA 175
EE WISWFC L G++ F EVDE+YI+D FNL GL V ++ A+DMIL PDEE
Sbjct 5 EEVSWISWFCGLRGNEFFCEVDEDYIQDKFNLTGLNEQVPHYRQALDMILDLEPDEELED 64
Query 176 DQSFMDIYRDAVD-LYGLIHARYIISPRGLPQMRDRCIDEIFGHCPRVLCDRQVLLPVGI 234
+ + D+ A + LYGLIHARYI++ RG+ QM ++ FG+CPRV C+ Q +LP+G+
Sbjct 65 NPNQSDLIEQAAEMLYGLIHARYILTNRGIAQMLEKYQQGDFGYCPRVYCENQPMLPIGL 124
Query 235 SPDLRLHKLQLFCPLCQEVYDMVNAHERTKDIDGAYFGPSFPHIFLQTYSSWVPSVAPIP 294
S ++L+CP C +VY ++ R DGAYFG FPH+ + + P
Sbjct 125 SDIPGEAMVKLYCPKCMDVYTPKSS--RHHHTDGAYFGTGFPHMLFMVHPEYRPKRPANQ 182
Query 295 YQPRLFGFRIH 305
+ PRL+GF+IH
Sbjct 183 FVPRLYGFKIH 193
> At4g17640
Length=282
Score = 166 bits (421), Expect = 6e-41, Method: Compositional matrix adjust.
Identities = 86/208 (41%), Positives = 126/208 (60%), Gaps = 9/208 (4%)
Query 102 EEEEEEEEEEEEEEEEEGWISWFCSLEGHDCFAEVDEEYIRDTFNLFGLKSAVNNFEGAM 161
+ + EE E + E+ WISWFC+L G++ F EVD++YI+D FNL GL V ++ A+
Sbjct 78 DTDSEESEVSGSDGEDTSWISWFCNLRGNEFFCEVDDDYIQDDFNLCGLSHQVPYYDYAL 137
Query 162 DMILGDAPDEEEVADQSFMDIYRDAVD-LYGLIHARYIISPRGLPQMRDRCIDEIFGHCP 220
D+IL E+ + ++ A + LYG+IHAR+I++ +GL M D+ + FG CP
Sbjct 138 DLILDVESSHGEMFTEEQNELIESAAEMLYGMIHARFILTSKGLASMLDKYKNYDFGRCP 197
Query 221 RVLCDRQVLLPVGISPDLRLHKLQLFCPLCQEVYDMVNAHERTK---DIDGAYFGPSFPH 277
RV C Q LPVG S R ++++CP C++VY + R+K +IDGAYFG +FPH
Sbjct 198 RVYCCGQPCLPVGQSDIPRASTVKIYCPKCEDVY-----YPRSKYQGNIDGAYFGTTFPH 252
Query 278 IFLQTYSSWVPSVAPIPYQPRLFGFRIH 305
+FL TY P A Y R+FGF++H
Sbjct 253 LFLMTYGHLKPQKASQSYTQRVFGFKLH 280
> At2g44680
Length=283
Score = 164 bits (416), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 82/213 (38%), Positives = 127/213 (59%), Gaps = 9/213 (4%)
Query 100 EEEEEEEEEEEEEEEEEEEGWISWFCSLEGHDCFAEVDEEYIRDTFNLFGLKSAVNNFEG 159
E + + E + E ++ WISWFC+L G++ F EVDE+YI+D FNL GL V ++
Sbjct 76 ESDTDSEGSDVSGSEGDDTSWISWFCNLRGNEFFCEVDEDYIQDDFNLCGLSGQVPYYDY 135
Query 160 AMDMILGDAPDEEEVADQSFMDIYRDAVD-LYGLIHARYIISPRGLPQMRDRCIDEIFGH 218
A+D+IL ++ + ++ A + LYGLIH RYI++ +G+ M ++ + FG
Sbjct 136 ALDLILDVESSNGDMFTEEQHEMVESAAEMLYGLIHVRYILTTKGMAAMMEKYKNYDFGR 195
Query 219 CPRVLCDRQVLLPVGISPDLRLHKLQLFCPLCQEVYDMVNAHERTK---DIDGAYFGPSF 275
CPRV C Q LPVG S R ++++CP C+++Y + R+K +IDGAYFG +F
Sbjct 196 CPRVFCCGQSCLPVGQSDIPRSSTVKIYCPKCEDIY-----YPRSKYQGNIDGAYFGTTF 250
Query 276 PHIFLQTYSSWVPSVAPIPYQPRLFGFRIHNRH 308
PH+FL Y + P Y P++FGF++HN+
Sbjct 251 PHLFLMAYGNMKPQKPAQNYVPKIFGFKVHNKQ 283
> 7292695
Length=215
Score = 163 bits (412), Expect = 6e-40, Method: Compositional matrix adjust.
Identities = 83/200 (41%), Positives = 121/200 (60%), Gaps = 3/200 (1%)
Query 116 EEEGWISWFCSLEGHDCFAEVDEEYIRDTFNLFGLKSAVNNFEGAMDMILGDAPDEEEVA 175
EE W++WFC L G++ F EVDE+YI+D FNL GL V N+ A+DMIL P++E
Sbjct 5 EEVSWVTWFCGLRGNEFFCEVDEDYIQDKFNLTGLNEQVPNYRQALDMILDLEPEDELED 64
Query 176 DQSFMDIYRDAVD-LYGLIHARYIISPRGLPQMRDRCIDEIFGHCPRVLCDRQVLLPVGI 234
+ D+ A + LYGLIHARYI++ RG+ QM ++ FGHCPRV C+ Q +LP+G+
Sbjct 65 NPLQSDMTEQAAEMLYGLIHARYILTNRGIAQMIEKYQTGDFGHCPRVYCESQPMLPLGL 124
Query 235 SPDLRLHKLQLFCPLCQEVYDMVNAHERTKDIDGAYFGPSFPHIFLQTYSSWVPSVAPIP 294
S ++ +CP C +VY ++ R DGAYFG FPH+ + + P
Sbjct 125 SDIPGEAMVKTYCPKCIDVYTPKSS--RHHHTDGAYFGTGFPHMLFMVHPEYRPKRPTNQ 182
Query 295 YQPRLFGFRIHNRHSIIKIK 314
+ PRL+GF+IH+ I+++
Sbjct 183 FVPRLYGFKIHSLAYQIQLQ 202
> CE06343
Length=235
Score = 162 bits (409), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 81/191 (42%), Positives = 116/191 (60%), Gaps = 3/191 (1%)
Query 116 EEEGWISWFCSLEGHDCFAEVDEEYIRDTFNLFGLKSAVNNFEGAMDMILGDAPDEEEVA 175
EE WI+WFC L G++ F EVDEEYI+D FNL GL V + A+DMIL P+++
Sbjct 5 EEVSWITWFCGLRGNEFFCEVDEEYIQDRFNLTGLNEQVPKYRQALDMILDLEPEDDIED 64
Query 176 DQSFMDIYRDAVD-LYGLIHARYIISPRGLPQMRDRCIDEIFGHCPRVLCDRQVLLPVGI 234
+ + D+ A + LYGLIHARYI++ RG+ QM ++ D FG CPRV C+ Q +LP+G+
Sbjct 65 NATNTDLVEQAAEMLYGLIHARYILTNRGISQMVEKWRDHDFGVCPRVYCENQPMLPIGL 124
Query 235 SPDLRLHKLQLFCPLCQEVYDMVNAHERTKDIDGAYFGPSFPHIFLQTYSSWVPSVAPIP 294
S ++L+CP C +V+ V R + DG+YFG FPH+ + P
Sbjct 125 SDVPGEAMVKLYCPRCNDVF--VPRSSRHQHTDGSYFGTGFPHMLFFVHPDLRPRRPVTQ 182
Query 295 YQPRLFGFRIH 305
+ P+L+GF+IH
Sbjct 183 FVPKLYGFKIH 193
> SPAC1851.03
Length=231
Score = 160 bits (406), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 83/195 (42%), Positives = 119/195 (61%), Gaps = 8/195 (4%)
Query 120 WISWFCSLEGHDCFAEVDEEYIRDTFNLFGLKSAVNNFEGAMDMILG--DAPDEEEVADQ 177
W+ WF L+G++ F EVDE++I+D FNL GL V ++ ++D+IL D EEV D
Sbjct 16 WVDWFLGLKGNEFFCEVDEDFIQDRFNLTGLSHEVPHYSQSLDLILDVLDPDLPEEVQD- 74
Query 178 SFMDIYRDAVDLYGLIHARYIISPRGLPQMRDRCIDEIFGHCPRVLCDRQVLLPVGISPD 237
++ A LYGLIHARYI++ +GL +M ++ FGHCPRVLC+ Q +LPVG+S
Sbjct 75 ---EVEASARHLYGLIHARYILTAQGLYKMLEKYKKCDFGHCPRVLCNGQPMLPVGLSDI 131
Query 238 LRLHKLQLFCPLCQEVYDMVNAHERTKDIDGAYFGPSFPHIFLQTYSSWVPSVAPIPYQP 297
++L+CP C++VY +R IDGAYFG SFPH+ Q Y + Y P
Sbjct 132 AHTKSVKLYCPRCEDVY--TPKSQRHASIDGAYFGTSFPHMLFQVYPELAVPKSQERYIP 189
Query 298 RLFGFRIHNRHSIIK 312
R+FGF++H+ + K
Sbjct 190 RIFGFKVHSYSATFK 204
> CE18168
Length=234
Score = 160 bits (406), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 83/191 (43%), Positives = 116/191 (60%), Gaps = 4/191 (2%)
Query 116 EEEGWISWFCSLEGHDCFAEVDEEYIRDTFNLFGLKSAVNNFEGAMDMILGDAPDEEEVA 175
EE WI+WFC L G++ F EVDEEYI+D FNL GL V + A+DMIL PD+ E
Sbjct 5 EEVSWITWFCGLRGNEFFCEVDEEYIQDRFNLTGLNEQVPKYRQALDMILDLEPDDIE-D 63
Query 176 DQSFMDIYRDAVD-LYGLIHARYIISPRGLPQMRDRCIDEIFGHCPRVLCDRQVLLPVGI 234
+ + D+ A + LYGLIHARYI++ RG+ QM ++ D FG CPRV C+ Q +LP+G+
Sbjct 64 NATNTDLVEQAAEMLYGLIHARYILTNRGISQMVEKWRDHDFGVCPRVYCENQPMLPIGL 123
Query 235 SPDLRLHKLQLFCPLCQEVYDMVNAHERTKDIDGAYFGPSFPHIFLQTYSSWVPSVAPIP 294
S ++L+CP C +V+ V R + DG+YFG FPH+ + P
Sbjct 124 SDVPGEAMVKLYCPRCNDVF--VPRSSRHQHTDGSYFGTGFPHMLFFVHPDLRPRRPVTQ 181
Query 295 YQPRLFGFRIH 305
+ P+L+GF+IH
Sbjct 182 FVPKLYGFKIH 192
> 7302395
Length=219
Score = 150 bits (379), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 77/192 (40%), Positives = 112/192 (58%), Gaps = 3/192 (1%)
Query 114 EEEEEGWISWFCSLEGHDCFAEVDEEYIRDTFNLFGLKSAVNNFEGAMDMILGDAPDEEE 173
+ +E WI WFC G++ F EVDEEYI+D FNL L S V N++ A+++IL P
Sbjct 3 DSDESSWIHWFCKQRGNEFFCEVDEEYIQDKFNLNFLDSNVKNYKCALEVILDLNPGSAS 62
Query 174 VADQSFMDIYRDAVDLYGLIHARYIISPRGLPQMRDRCIDEIFGHCPRVLCDRQVLLPVG 233
D + ++ A LYGLIHAR+I++ RG+ M D+ FG CPR C Q +LP+G
Sbjct 63 -EDPAEPELEASAEKLYGLIHARFILTNRGIELMLDKYNKGEFGTCPRAFCHSQPVLPIG 121
Query 234 ISPDLRLHKLQLFCPLCQEVYDMVNAHERTKDIDGAYFGPSFPHIFLQTYSSWVPSVAPI 293
+S + ++++CP C +VY + R ++DGA+FG FPH+F P A
Sbjct 122 LSDNPGEDMVRIYCPKCNDVY--IPKASRHSNLDGAFFGTGFPHMFFMEKPDARPKRAKQ 179
Query 294 PYQPRLFGFRIH 305
+ PRL+GF+IH
Sbjct 180 KFVPRLYGFKIH 191
> YOR039w
Length=258
Score = 150 bits (378), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 77/195 (39%), Positives = 110/195 (56%), Gaps = 4/195 (2%)
Query 120 WISWFCSLEGHDCFAEVDEEYIRDTFNLFGLKSAVNNFEGAMDMILGDAPDE--EEVADQ 177
WI F +GH+ F +VD EYI D FNL L+ V+ F + I+ D D E +
Sbjct 38 WIDLFLGRKGHEYFCDVDPEYITDRFNLMNLQKTVSKFSYVVQYIVDDLDDSILENMTHA 97
Query 178 SFMDIYRDAVDLYGLIHARYIISPRGLPQMRDRCIDEIFGHCPRVLCDRQVLLPVGISPD 237
+ D+ LYGLIHARYII+ +GL +M + + FG CPRV C+ Q LLPVG+
Sbjct 98 RLEQLESDSRKLYGLIHARYIITIKGLQKMYAKYKEADFGRCPRVYCNLQQLLPVGLHDI 157
Query 238 LRLHKLQLFCPLCQEVYDMVNAHERTKDIDGAYFGPSFPHIFLQTYSSWVPSVAPIPYQP 297
+ ++L+CP C+++Y + R IDGAYFG SFP +FLQ + VP Y P
Sbjct 158 PGIDCVKLYCPSCEDLY--IPKSSRHSSIDGAYFGTSFPGMFLQAFPDMVPKHPTKRYVP 215
Query 298 RLFGFRIHNRHSIIK 312
++FGF +H + + +
Sbjct 216 KIFGFELHKQAQLTR 230
> SPBC2G5.02c
Length=254
Score = 149 bits (375), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 86/210 (40%), Positives = 121/210 (57%), Gaps = 10/210 (4%)
Query 100 EEEEEEEEEEEEEEEEEEEGWISWFCSLEGHDCFAEVDEEYIRDTFNLFGLKSAVNNFEG 159
++ +E ++ E WISWFCS G + F EV E++I D FNL GL AV +
Sbjct 21 DDADELFDDLSSSPLHENVSWISWFCSRPGREYFVEVKEDFIEDLFNLTGLNLAVPFYNE 80
Query 160 AMDMILG-DAPDEEEVADQSFMDIYRDAVDLYGLIHARYIISPRGLPQMRDRCIDEIFGH 218
A+D+IL APD E D ++ A LYGLIH RYII+ GL QM ++ IFG
Sbjct 81 ALDLILDRTAPDTLENFDMDVIET--SAQILYGLIHQRYIITRTGLHQMAEKYSMGIFGC 138
Query 219 CPRVLCDRQVLLPVGISPDLRLHKLQLFCPLCQEVYDMVNAHERTKDIDGAYFGPSFPHI 278
CPRV C +LP G+S + + LFCP C ++Y + R K+IDG++FG +FPH+
Sbjct 139 CPRVNCCYTHVLPAGLSDIVGKMPVMLFCPNCLDLY--APSSSRYKNIDGSFFGATFPHL 196
Query 279 FLQTYSSWVPSVAPIP----YQPRLFGFRI 304
F ++Y P + IP YQPR++GF++
Sbjct 197 FFESYPELNPKRS-IPCGKIYQPRIYGFKV 225
> 7291794
Length=219
Score = 130 bits (328), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 71/189 (37%), Positives = 103/189 (54%), Gaps = 14/189 (7%)
Query 119 GWISWFCSLEGHDCFAEVDEEYIRDTFNLFGLKSAVNNFEGAMDMILGDAPDEEEVADQS 178
WI WF ++GH+ V EYI+D FNL GL+ E +D + ++ + A++
Sbjct 13 SWIDWFLGIKGHEFSCRVPNEYIQDKFNLTGLEFDSQTLEVVLDPEFDN--EDWDCAEEK 70
Query 179 FMDIYRDAVDLYGLIHARYIISPRGLPQMRDRCIDEIFGHCPRVLCDRQVLLPVGISPDL 238
LYG+IHARYI+SPRG+ MR + FG CPRV C RQ +LPVG+
Sbjct 71 ---------KLYGMIHARYIVSPRGIEDMRLKYERGDFGSCPRVFCKRQKVLPVGLHDVW 121
Query 239 RLHKLQLFCPLCQEVYDMVNAHERTKDIDGAYFGPSFPHIFLQTYSSWVPSVAPIPYQPR 298
+++++CP C VY + +DGA FG SFPH+F S +PS Y PR
Sbjct 122 DKAQVKIYCPSCNNVYIPL---PHNGMLDGAMFGTSFPHMFFMQLPSLIPSPPVEKYIPR 178
Query 299 LFGFRIHNR 307
++GF++H +
Sbjct 179 IYGFQLHKK 187
> YGL019w
Length=278
Score = 116 bits (290), Expect = 8e-26, Method: Compositional matrix adjust.
Identities = 70/223 (31%), Positives = 113/223 (50%), Gaps = 40/223 (17%)
Query 120 WISWFCSLEGHDCFAEVDEEYIRDTFNLFGLKSAVNNFEGAMDMIL-------------- 165
WI FCS GH+ F +V E+I D FN+ L V ++ A+D+IL
Sbjct 26 WIPSFCSRFGHEYFCQVPTEFIEDDFNMTSLSQEVPHYRKALDLILDLEAMSDEEEDEDD 85
Query 166 -------------------GDAPDEEEVADQSFMDIYRDAVDLYGLIHARYIISPRGLPQ 206
G ++ V ++S ++ A LYGLIHAR+I++ GL
Sbjct 86 VVEEDEVDQEMQSNDGHDEGKRRNKSPVVNKSIIE--HAAEQLYGLIHARFILTKPGLQA 143
Query 207 MRDRCIDEIFGHCPRVLCDRQVLLPVGISPDLRLHKLQLFCPLCQEVYDMVNAHERTKDI 266
M ++ + FG CPR C+ LLP G+S + H ++L+CP CQ++Y + R +
Sbjct 144 MAEKFDHKEFGTCPRYYCNGMQLLPCGLSDTVGKHTVRLYCPSCQDLY--LPQSSRFLCL 201
Query 267 DGAYFGPSFPHIFLQTYS---SWVPSVAPIPYQPRLFGFRIHN 306
+GA++G SFP +FL+ + +V + Y+ ++FGFRI++
Sbjct 202 EGAFWGTSFPGVFLKHFKELEEYVERKSKESYELKVFGFRIND 244
> ECU10g1200i
Length=205
Score = 100 bits (250), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 58/200 (29%), Positives = 96/200 (48%), Gaps = 13/200 (6%)
Query 105 EEEEEEEEEEEEEEGWISWFCSLEGHDCFAEVDEEYIRDTFNLFGLKSAVNNFEGAMDMI 164
E++ E +E W+ F + + V +E++ D FN GL ++N E + +
Sbjct 6 EDDSENLLRISSDEYWVGDFMRRKENHSIVRVPDEFLEDRFNTIGLDRYIDNLEEVYNSV 65
Query 165 LGDAPDEEEVADQSFMDIYRDAVDLYGLIHARYIISPRGLPQMRDRCIDEIFGHCPRVLC 224
L P +++ LY LIH RYI + GL + D+ + +G CPRV C
Sbjct 66 LDKGPRGN----------FKEESSLYYLIHQRYIFTKSGLEAVLDKVMSREYGACPRVGC 115
Query 225 DRQVLLPVGISPDLRLHKLQLFCPLCQEVYDMVNAHERTKDIDGAYFGPSFPHIFLQTYS 284
++P+G+S ++ +++C C VY+ A + +DG +G SFPH + T+S
Sbjct 116 RSVGVIPIGLSDRPQVSSTKIYCHSCVNVYE---ASGTLQLLDGCAWGRSFPHFLILTHS 172
Query 285 SWVPSVAPIPYQPRLFGFRI 304
PS Y PR++G RI
Sbjct 173 YHFPSRTCEEYIPRIYGLRI 192
Lambda K H
0.319 0.138 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8410937234
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40