bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2752_orf6
Length=417
Score E
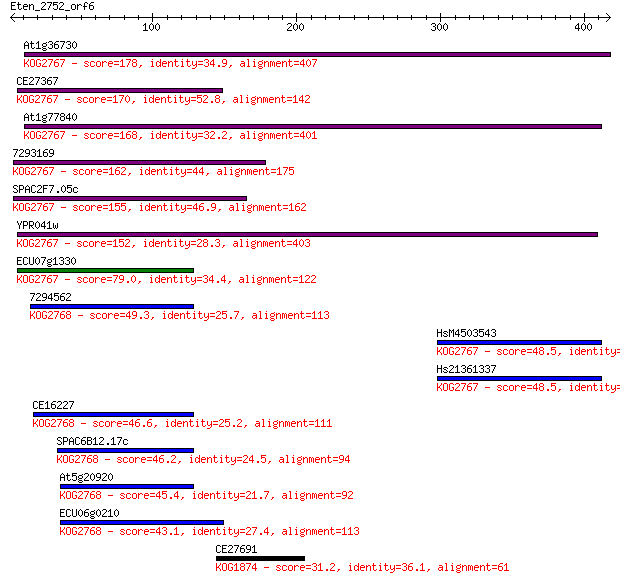
Sequences producing significant alignments: (Bits) Value
At1g36730 178 2e-44
CE27367 170 7e-42
At1g77840 168 3e-41
7293169 162 1e-39
SPAC2F7.05c 155 2e-37
YPR041w 152 1e-36
ECU07g1330 79.0 2e-14
7294562 49.3 1e-05
HsM4503543 48.5 3e-05
Hs21361337 48.5 3e-05
CE16227 46.6 9e-05
SPAC6B12.17c 46.2 1e-04
At5g20920 45.4 2e-04
ECU06g0210 43.1 0.001
CE27691 31.2 5.0
> At1g36730
Length=439
Score = 178 bits (451), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 142/438 (32%), Positives = 219/438 (50%), Gaps = 40/438 (9%)
Query 11 DREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKF 70
+R+D YRYKMP++++KIEGRGNGI+TNV NM EIA+AL RP Y TK+FGCELGA KF
Sbjct 10 NRDDAFYRYKMPRMMTKIEGRGNGIKTNVVNMVEIAKALGRPAAYTTKYFGCELGAQSKF 69
Query 71 EENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITVKKG-VLVCKCNACGYS 129
+E +L+NGAH L +L+ FI+ YV C GC PE +I + K +L KC ACG+
Sbjct 70 DEKNGTSLVNGAHDTSKLAGLLENFIKKYVQCYGCGNPETEILITKTQMLQLKCAACGFL 129
Query 130 GTLDNTHKAATYMAKNPPNATETTLGK---KKKTKEERRAEKHAKEEDKSEKTGKEKKSK 186
+D K +++ KNPP +++ K ++ KE R + A EE + K K K
Sbjct 130 SDVDMRDKLTSFILKNPPEQKKSSKDKKSMRRAEKERLREGEAADEEMRKLKKEAASKKK 189
Query 187 DKDKKKKKGEEENDTDCLNSKAVDHSGDATPTNAGED--SWDEEKSSEADKKEKKKK--- 241
K + D +++ D + +D W + S EA +K K++
Sbjct 190 AATTGTSKDKVSKKKDHSPPRSLSDENDQADSEEDDDDVQWQTDTSREAAEKRMKEQLSA 249
Query 242 -----------EKKHANAEGPKL--VVKEALTINCPEIAEVAARLRNMI--VTSSAVTPD 286
E+K AE K V E PE A +L N I + SS +P
Sbjct 250 VTAEMVMLSTVEEKKPVAEVKKAPEQVHENGNSKIPENAH--EKLVNEIKELLSSGSSP- 306
Query 287 SFFVELRMLQVSQDFDAKCRMYVVLSALFKD---GLSPELLEEK---MAFVAKCCDSSVH 340
+L+ S + + +M + SALF G + E++++K +A + ++
Sbjct 307 ---TQLKTALASNSANPQEKMDALFSALFGGTGKGFAKEVIKKKKYLLALMMMQEEAGAP 363
Query 341 AS-DLLSALENFCFENPHTAMAGYPYMLQRLYNAELLEPEDILAYYNSPRADAATQKCRT 399
A LL+ +E+FC + A +++ LY+ ++L+ + I+ +YN + K T
Sbjct 364 AQMGLLNGIESFCMKASAEAAKEVALVIKGLYDEDILDEDVIVEWYNKGVKSSPVLKNVT 423
Query 400 FAEPFLKWLAEADSSDEE 417
PF++WL A+S EE
Sbjct 424 ---PFIEWLQNAESESEE 438
> CE27367
Length=436
Score = 170 bits (430), Expect = 7e-42, Method: Compositional matrix adjust.
Identities = 75/142 (52%), Positives = 99/142 (69%), Gaps = 0/142 (0%)
Query 6 VNIPRDREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELG 65
+N+ R DP YRYKMPKL +K+EG+GNGI+T + NM EIA+AL+RPPMYPTK+FGCELG
Sbjct 3 LNVNRAVADPFYRYKMPKLSAKVEGKGNGIKTVISNMSEIAKALERPPMYPTKYFGCELG 62
Query 66 AMVKFEENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITVKKGVLVCKCNA 125
A F+ E+ ++NG H L ILD FI+ +VLC C+ PE + V+K + KC A
Sbjct 63 AQTNFDAKNERYIVNGEHDANKLQDILDGFIKKFVLCKSCENPETQLFVRKNNIKSKCKA 122
Query 126 CGYSGTLDNTHKAATYMAKNPP 147
CG S +D HK +T++ KNPP
Sbjct 123 CGCSFDIDLKHKLSTFIMKNPP 144
> At1g77840
Length=437
Score = 168 bits (425), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 129/434 (29%), Positives = 211/434 (48%), Gaps = 47/434 (10%)
Query 11 DREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKF 70
+R D YRYKMPK+++K EG+GNGI+TN+ N EIA+AL RPP Y TK+FGCELGA KF
Sbjct 10 NRNDAFYRYKMPKMVTKTEGKGNGIKTNIINNVEIAKALARPPSYTTKYFGCELGAQSKF 69
Query 71 EENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITVKKGVLV-CKCNACGYS 129
+E +L+NGAH+ L +L+ FI+ +V C GC PE +I + K +V KC ACG+
Sbjct 70 DEKTGTSLVNGAHNTSKLAGLLENFIKKFVQCYGCGNPETEIIITKTQMVNLKCAACGFI 129
Query 130 GTLDNTHKAATYMAKNPPNATETTLGKK--KKTKEERRAEKH-AKEEDKSEKTGKEKKSK 186
+D K ++ KNPP + + KK +K ++ER E A EE + K K+ S
Sbjct 130 SEVDMRDKLTNFILKNPPEQKKVSKDKKAMRKAEKERLKEGELADEEQRKLKAKKKALSN 189
Query 187 DKDKKKKKGEEENDTDCLNSKAVDHSGDATPTNAGEDSWDEEKSSEADKKEKKKKEKKHA 246
KD K K ++ D + W + S EA +K+ +
Sbjct 190 GKDSKTSKNHSSDEDISPKHDENALEVDEDEDDDDGVEWQTDTSREA----AEKRMMEQL 245
Query 247 NAEGPKLVVKEALTIN-------------------CPEIAEVAARLRNMIVTSSAVTPDS 287
+A+ ++V+ A+ + P+ + ++ + S ++
Sbjct 246 SAKTAEMVMLSAMEVEEKKAPKSKSNGNVVKTENPPPQEKNLVQDMKEYLKKGSPISALK 305
Query 288 FFVELRMLQVSQDFDAKCRMYVVLSALFKD---GLSPELLEEKMAFVAKCC----DSSVH 340
F+ + + QD M + +ALF G + E+ ++K A S +H
Sbjct 306 SFIS-SLSEPPQDI-----MDALFNALFDGVGKGFAKEVTKKKNYLAAAATMQEDGSQMH 359
Query 341 ASDLLSALENFCFENPH-TAMAGYPYMLQRLYNAELLEPEDILAYYNS--PRADAATQKC 397
LL+++ FC +N + A+ +L+ LY+ +++E E +L +Y AD ++
Sbjct 360 ---LLNSIGTFCGKNGNEEALKEVALVLKALYDQDIIEEEVVLDWYEKGLTGADKSSPVW 416
Query 398 RTFAEPFLKWLAEA 411
+ +PF++WL A
Sbjct 417 KN-VKPFVEWLQSA 429
> 7293169
Length=464
Score = 162 bits (410), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 77/177 (43%), Positives = 108/177 (61%), Gaps = 4/177 (2%)
Query 3 MQYVNIPRDREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGC 62
M VN+ R D YRYKMP+L +K+EG+GNGI+T + NM E+ARA+ RP YPTK+FGC
Sbjct 1 MATVNVNRSVTDIFYRYKMPRLQAKVEGKGNGIKTVLVNMAEVARAIGRPATYPTKYFGC 60
Query 63 ELGAMVKFEENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITV--KKGVLV 120
ELGA F+ E+ ++NG+H L +LD FI+ +VLC CD PE ++TV K +
Sbjct 61 ELGAQTLFDHKNERFVVNGSHDVNKLQDLLDGFIRKFVLCPECDNPETNLTVSAKNQTIS 120
Query 121 CKCNACGYSGTLDNTHKAATYMAKNPPNATETTLGKKKKTKEERRAEKHAKEEDKSE 177
C ACG+ G L HK T++ KNPP+ G E +R+ K ++ D ++
Sbjct 121 QSCKACGFHGLLKVNHKVNTFIVKNPPSLNPAAQG--SSLTEGKRSRKQKQKNDNAD 175
> SPAC2F7.05c
Length=395
Score = 155 bits (392), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 76/164 (46%), Positives = 101/164 (61%), Gaps = 9/164 (5%)
Query 3 MQYVNIPRDREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGC 62
M +NI RD +D YRY+MPKL SKIEG+GNGI+T + NM +IA+AL RPP+Y TKFFG
Sbjct 1 MATINIRRDVKDSFYRYRMPKLQSKIEGKGNGIKTVIPNMSDIAKALGRPPLYVTKFFGF 60
Query 63 ELGAMVKFEENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITVKK--GVLV 120
ELGA + ++ ++NGAH L +LD FI+ +VLC C PE ++++ K +
Sbjct 61 ELGAQTTIIADMDRYIVNGAHDAGKLQDLLDVFIRRFVLCASCQNPETELSINKKDQTIS 120
Query 121 CKCNACGYSGTLDNTHKAATYMAKNPPNATETTLGKKKKTKEER 164
C ACGY G +D HK + KNPP KKK K +R
Sbjct 121 YDCKACGYRGVIDGRHKLTGVIVKNPP-------AKKKSHKHKR 157
> YPR041w
Length=405
Score = 152 bits (384), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 114/410 (27%), Positives = 177/410 (43%), Gaps = 27/410 (6%)
Query 6 VNIPRDREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELG 65
+NI RD DP YRYKMP + +K+EGRGNGI+T V N+ +I+ AL RP Y K+FG ELG
Sbjct 3 INICRDNHDPFYRYKMPPIQAKVEGRGNGIKTAVLNVADISHALNRPAPYIVKYFGFELG 62
Query 66 AMVKFEENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITVKK-GVLVCKCN 124
A ++++ L+NG H L +LD FI +VLCG C PE +I + K LV C
Sbjct 63 AQTSISVDKDRYLVNGVHEPAKLQDVLDGFINKFVLCGSCKNPETEIIITKDNDLVRDCK 122
Query 125 ACGYSGTLDNTHKAATYMAKNPPNATETTLGKKKKTKEERRAEKHAKEEDKSEKTGKEKK 184
ACG +D HK ++++ KNPP++ + KKK + +
Sbjct 123 ACGKRTPMDLRHKLSSFILKNPPDSVSGSKKKKKAATASANVRGGGLSISDIAQGKSQNA 182
Query 185 SKDKDKKKKKGEEENDTDCLNSKAVDHSGDATPTNAGEDSWDEEKSSEADKKEKKKKEKK 244
D + D D L+ + + +D W + S EA + K+ E
Sbjct 183 PSDGTGSSTPQHHDEDEDELSRQIKAAASTLEDIEVKDDEWAVDMSEEAIRARAKELE-- 240
Query 245 HANAEGPKLVVKEALTINCPEIAEVAARLRNMIVTSSAVTPDS--FFVELRMLQVSQDFD 302
V LT ++ E + P + + L V D
Sbjct 241 ----------VNSELT----QLDEYGEWILEQAGEDKENLPSDVELYKKAAELDVLNDPK 286
Query 303 AKCRMYVVLSALFKDGLSPELLEEKMAFVAKCCDSSVHASDLLSALENFCFENPHTAMAG 362
C V+ LF + + E+ E AF K + + + + +E F +
Sbjct 287 IGC---VLAQCLFDEDIVNEIAEHN-AFFTKILVTPEYEKNFMGGIERFLGLEHKDLIPL 342
Query 363 YPYMLQRLYNAELLEPEDILAYYNSPR----ADAATQKCRTFAEPFLKWL 408
P +L +LYN +++ E+I+ + ++K R A+PF+ WL
Sbjct 343 LPKILVQLYNNDIISEEEIMRFGTKSSKKFVPKEVSKKVRRAAKPFITWL 392
> ECU07g1330
Length=281
Score = 79.0 bits (193), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 42/122 (34%), Positives = 64/122 (52%), Gaps = 3/122 (2%)
Query 6 VNIPRDREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELG 65
++I R D YRYKMP+ + K EG+ RT + N+ EI +LKRPP+Y KF EL
Sbjct 2 IDINRTNNDFYYRYKMPRAVVKQEGKAGNTRTLIVNLEEIGSSLKRPPLYILKFMSYELA 61
Query 66 AMVKFEENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITVKKGVLVCKCNA 125
+ + + + +NG + + ++ FI +V+C C PE + G L +C A
Sbjct 62 T--RTDVGKGRYAVNGKYESSRIQDLIYNFIDEFVICPFCSNPET-FYINSGGLSMECLA 118
Query 126 CG 127
CG
Sbjct 119 CG 120
> 7294562
Length=312
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/113 (25%), Positives = 52/113 (46%), Gaps = 6/113 (5%)
Query 15 PNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKFEENE 74
P + + P++L R +T+ N +IA+ L R P + F ELG + N+
Sbjct 179 PKFVMRPPQVL-----RVGTKKTSFANFMDIAKTLHRLPKHLLDFLLAELGTSGSMDGNQ 233
Query 75 EKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITVKKGVLVCKCNACG 127
+ +I G + + +L ++I+ YV C C PE + + +C +CG
Sbjct 234 Q-LIIKGRFQPKQIENVLRRYIKEYVTCHTCRSPETILQKDTRLFFLQCESCG 285
> HsM4503543
Length=431
Score = 48.5 bits (114), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 34/121 (28%), Positives = 62/121 (51%), Gaps = 9/121 (7%)
Query 298 SQDFDAKCRMYVVLS-ALFKDGLSPELLEEKMAFVAKCCDSSVHASDLLSALENFCFENP 356
++ D K +VL+ LF + + ++ + + F+ C ++ LL LE C
Sbjct 267 AERLDVKAMGPLVLTEVLFNEKIREQIKKYRRHFLRFCHNNKKAQRYLLHGLE--CVVAM 324
Query 357 HTA--MAGYPYMLQRLYNAELLEPEDILAYYNSPRADAAT----QKCRTFAEPFLKWLAE 410
H A ++ P++L+ +Y+A+LLE E I+++ + ++ R AEPF+KWL E
Sbjct 325 HQAQLISKIPHILKEMYDADLLEEEVIISWSEKASKKYVSKELAKEIRVKAEPFIKWLKE 384
Query 411 A 411
A
Sbjct 385 A 385
> Hs21361337
Length=431
Score = 48.5 bits (114), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 34/121 (28%), Positives = 62/121 (51%), Gaps = 9/121 (7%)
Query 298 SQDFDAKCRMYVVLS-ALFKDGLSPELLEEKMAFVAKCCDSSVHASDLLSALENFCFENP 356
++ D K +VL+ LF + + ++ + + F+ C ++ LL LE C
Sbjct 267 AERLDVKAMGPLVLTEVLFNEKIREQIKKYRRHFLRFCHNNKKAQRYLLHGLE--CVVAM 324
Query 357 HTA--MAGYPYMLQRLYNAELLEPEDILAYYNSPRADAAT----QKCRTFAEPFLKWLAE 410
H A ++ P++L+ +Y+A+LLE E I+++ + ++ R AEPF+KWL E
Sbjct 325 HQAQLISKIPHILKEMYDADLLEEEVIISWSEKASKKYVSKELAKEIRVKAEPFIKWLKE 384
Query 411 A 411
A
Sbjct 385 A 385
> CE16227
Length=250
Score = 46.6 bits (109), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 28/111 (25%), Positives = 49/111 (44%), Gaps = 6/111 (5%)
Query 17 YRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKFEENEEK 76
+ K+P++ R +T N EI R +KR + +F ELG +
Sbjct 114 FAIKLPEV-----ARAGSKKTAFSNFLEICRLMKRQDKHVLQFLLAELGTTGSID-GSNC 167
Query 77 ALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITVKKGVLVCKCNACG 127
++ G ++ ++L K+I+ YV+C C PE +T + +C CG
Sbjct 168 LIVKGRWQQKQFESVLRKYIKEYVMCHTCKSPETQLTKDTRLFFLQCTNCG 218
> SPAC6B12.17c
Length=310
Score = 46.2 bits (108), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 23/94 (24%), Positives = 44/94 (46%), Gaps = 1/94 (1%)
Query 34 GIRTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKFEENEEKALINGAHSEEDLVAILD 93
G +T N+ +I++ + R + +F ELG + + +I G ++ + +L
Sbjct 195 GKKTIFANISDISKRMHRSLDHVIQFLFAELGTSGSVD-GSSRLIIKGRFQQKQIENVLR 253
Query 94 KFIQMYVLCGGCDLPEIDITVKKGVLVCKCNACG 127
++I YV C C P+ +T + + C ACG
Sbjct 254 RYIVEYVTCKTCKSPDTILTKENRIFFMTCEACG 287
> At5g20920
Length=268
Score = 45.4 bits (106), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/92 (21%), Positives = 46/92 (50%), Gaps = 1/92 (1%)
Query 36 RTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKFEENEEKALINGAHSEEDLVAILDKF 95
+T N ++ + + R P + ++ ELG + +++ ++ G + ++ IL ++
Sbjct 157 KTVFVNFMDLCKTMHRQPDHVMQYLLAELGTSGSLD-GQQRLVVKGRFAPKNFEGILRRY 215
Query 96 IQMYVLCGGCDLPEIDITVKKGVLVCKCNACG 127
I YV+C GC P+ ++ + + +C CG
Sbjct 216 ITDYVICLGCKSPDTILSKENRLFFLRCEKCG 247
> ECU06g0210
Length=228
Score = 43.1 bits (100), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 31/115 (26%), Positives = 53/115 (46%), Gaps = 4/115 (3%)
Query 36 RTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKFEENEE-KALINGAHSEEDLVAILDK 94
R N+ EI+R L R + F EL M N++ K L+ G ++ +L +
Sbjct 111 RKTSINIAEISRILNRSADHLVSFLCNEL--MTTGSINKDGKLLLKGMFIRSEIQEVLRR 168
Query 95 FIQMYVLCGGCD-LPEIDITVKKGVLVCKCNACGYSGTLDNTHKAATYMAKNPPN 148
+I+ +V+C C+ + + DI K + KC+ CG + + N + T K P
Sbjct 169 YIEHFVVCKICESVEDTDIVRDKRLYFLKCSKCGGARCVGNAVEGITTKGKAKPQ 223
> CE27691
Length=1437
Score = 31.2 bits (69), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 36/61 (59%), Gaps = 2/61 (3%)
Query 144 KNPPNATETTLGKKKKTKEERRAEKHAKEEDKSEKTGKEKKSKDKDKKKKKGEEENDTDC 203
K P TE+ G+ KK + ER +K +E + EK KE++ KDK++K++ GE + +
Sbjct 1286 KESPKDTESEKGEGKK-RRERSKDKETGKEKRLEKDNKERE-KDKERKRENGENQKEQKI 1343
Query 204 L 204
L
Sbjct 1344 L 1344
Lambda K H
0.313 0.129 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10699707996
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40