bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2753_orf2
Length=178
Score E
Sequences producing significant alignments: (Bits) Value
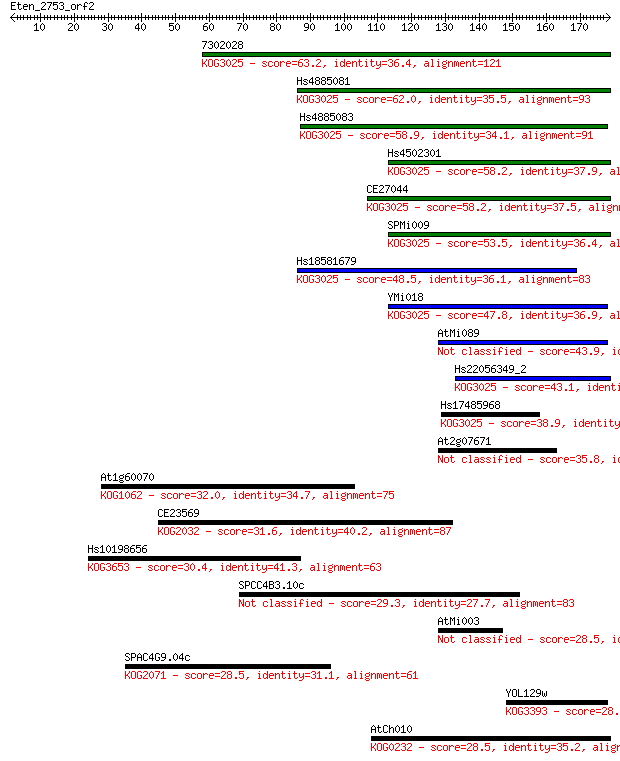
7302028 63.2 3e-10
Hs4885081 62.0 5e-10
Hs4885083 58.9 5e-09
Hs4502301 58.2 8e-09
CE27044 58.2 9e-09
SPMi009 53.5 2e-07
Hs18581679 48.5 7e-06
YMi018 47.8 1e-05
AtMi089 43.9 2e-04
Hs22056349_2 43.1 3e-04
Hs17485968 38.9 0.005
At2g07671 35.8 0.049
At1g60070 32.0 0.65
CE23569 31.6 0.85
Hs10198656 30.4 1.8
SPCC4B3.10c 29.3 4.5
AtMi003 28.5 7.1
SPAC4G9.04c 28.5 7.2
YOL129w 28.5 7.3
AtCh010 28.5 7.7
> 7302028
Length=138
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 44/122 (36%), Positives = 67/122 (54%), Gaps = 11/122 (9%)
Query 58 RALSTAPLLQRHSAVAQGPCARWPAAALTPLSSSSSSSSGGSPVGAVRHEASVAT-LSAA 116
R LS+A + Q + AQ P A L + S +S PV R S A + A
Sbjct 26 RPLSSAIISQSQTLAAQNTT---PVALLPQIRSFQTS-----PV--TRDIDSAAKFIGAG 75
Query 117 VALMSVGGVAQGIGSLFAALVSGTARNPSIKEDLFTYTLIGMGFLEFLGIICVMMSAVLL 176
A + V G GIG++F +L+ G ARNPS+K+ LF+Y ++G E +G+ C+MM+ +LL
Sbjct 76 AATVGVAGSGAGIGTVFGSLIIGYARNPSLKQQLFSYAILGFALSEAMGLFCLMMAFLLL 135
Query 177 YS 178
++
Sbjct 136 FA 137
> Hs4885081
Length=136
Score = 62.0 bits (149), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 33/104 (31%), Positives = 58/104 (55%), Gaps = 11/104 (10%)
Query 86 TPLSSSSSSSSGGSPVGAVRHEASVATLS-----------AAVALMSVGGVAQGIGSLFA 134
+P++SS S P+ R E + +S A A + V G GIG++F
Sbjct 32 SPVNSSKQPSYSNFPLQVARREFQTSVVSRDIDTAAKFIGAGAATVGVAGSGAGIGTVFG 91
Query 135 ALVSGTARNPSIKEDLFTYTLIGMGFLEFLGIICVMMSAVLLYS 178
+L+ G ARNPS+K+ LF+Y ++G E +G+ C+M++ ++L++
Sbjct 92 SLIIGYARNPSLKQQLFSYAILGFALSEAMGLFCLMVAFLILFA 135
> Hs4885083
Length=141
Score = 58.9 bits (141), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 31/91 (34%), Positives = 55/91 (60%), Gaps = 0/91 (0%)
Query 87 PLSSSSSSSSGGSPVGAVRHEASVATLSAAVALMSVGGVAQGIGSLFAALVSGTARNPSI 146
PL+S SS S + + + + + A A + V G GIG++F +L+ G ARNPS+
Sbjct 49 PLTSLVSSRSFQTSAISRDIDTAAKFIGAGAATVGVAGSGAGIGTVFGSLIIGYARNPSL 108
Query 147 KEDLFTYTLIGMGFLEFLGIICVMMSAVLLY 177
K+ LF+Y ++G E +G+ C+M++ ++L+
Sbjct 109 KQQLFSYAILGFALSEAMGLFCLMVAFLILF 139
> Hs4502301
Length=142
Score = 58.2 bits (139), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 25/66 (37%), Positives = 45/66 (68%), Gaps = 0/66 (0%)
Query 113 LSAAVALMSVGGVAQGIGSLFAALVSGTARNPSIKEDLFTYTLIGMGFLEFLGIICVMMS 172
+ A A + V G GIG++F +L+ G ARNPS+K+ LF+Y ++G E +G+ C+M++
Sbjct 76 IGAGAATVGVAGSGAGIGTVFGSLIIGYARNPSLKQQLFSYAILGFALSEAMGLFCLMVA 135
Query 173 AVLLYS 178
++L++
Sbjct 136 FLILFA 141
> CE27044
Length=116
Score = 58.2 bits (139), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 27/72 (37%), Positives = 46/72 (63%), Gaps = 0/72 (0%)
Query 107 EASVATLSAAVALMSVGGVAQGIGSLFAALVSGTARNPSIKEDLFTYTLIGMGFLEFLGI 166
+++ + A A + V G GIG++F ALV G ARNPS+K+ LF+Y ++G E +G+
Sbjct 44 DSAAKYIGAGAATVGVAGSGAGIGNVFGALVIGYARNPSLKQQLFSYAILGFALSEAMGL 103
Query 167 ICVMMSAVLLYS 178
C+ M ++L++
Sbjct 104 FCLTMGFMILFA 115
> SPMi009
Length=74
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 113 LSAAVALMSVGGVAQGIGSLFAALVSGTARNPSIKEDLFTYTLIGMGFLEFLGIICVMMS 172
+ A +A + V G GIG +F+ L+SGT+RNPS++ LF+ ++G E G+ C+M++
Sbjct 8 IGAGLATIGVSGAGVGIGLIFSNLISGTSRNPSVRPHLFSMAILGFALTEATGLFCLMLA 67
Query 173 AVLLYS 178
+++Y+
Sbjct 68 FLIIYA 73
> Hs18581679
Length=125
Score = 48.5 bits (114), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 30/94 (31%), Positives = 45/94 (47%), Gaps = 11/94 (11%)
Query 86 TPLSSSSSSSSGGSPVGAVRHEASVATLS-----------AAVALMSVGGVAQGIGSLFA 134
+P +S S SP+ R E + +S A A + V G GIG +F
Sbjct 32 SPDNSPKQPSYSSSPLRVARREFQTSIVSRDIDIAAKLIGAGAATVGVAGSGAGIGKVFG 91
Query 135 ALVSGTARNPSIKEDLFTYTLIGMGFLEFLGIIC 168
+L+ G ARN S+K+ LF+Y +G E +G+ C
Sbjct 92 SLIIGYARNLSLKQQLFSYATLGFALSEAMGLFC 125
> YMi018
Length=76
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 42/65 (64%), Gaps = 0/65 (0%)
Query 113 LSAAVALMSVGGVAQGIGSLFAALVSGTARNPSIKEDLFTYTLIGMGFLEFLGIICVMMS 172
+ A ++ + + G GI +FAAL++G +RNPSIK+ +F ++G E G+ C+M+S
Sbjct 10 IGAGISTIGLLGAGIGIAIVFAALINGVSRNPSIKDTVFPMAILGFALSEATGLFCLMVS 69
Query 173 AVLLY 177
+LL+
Sbjct 70 FLLLF 74
> AtMi089
Length=85
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 34/50 (68%), Gaps = 0/50 (0%)
Query 128 GIGSLFAALVSGTARNPSIKEDLFTYTLIGMGFLEFLGIICVMMSAVLLY 177
GIG++F++L+ ARNPS+ + LF Y ++G E + + +MM+ ++L+
Sbjct 34 GIGNVFSSLIHSVARNPSLAKQLFGYAILGFALTEAIALFALMMAFLILF 83
> Hs22056349_2
Length=126
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 32/46 (69%), Gaps = 0/46 (0%)
Query 133 FAALVSGTARNPSIKEDLFTYTLIGMGFLEFLGIICVMMSAVLLYS 178
F +L+ G ARN S+K+ LF Y ++G E +G+ C+M++ ++L++
Sbjct 80 FGSLIIGCARNLSLKQQLFFYAILGFALSEVMGLFCLMVAFLILFT 125
> Hs17485968
Length=124
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 15/29 (51%), Positives = 23/29 (79%), Gaps = 0/29 (0%)
Query 129 IGSLFAALVSGTARNPSIKEDLFTYTLIG 157
IG++F +L+ G ARNPS+K+ LF+Y +G
Sbjct 93 IGTVFGSLIIGYARNPSLKQQLFSYATLG 121
> At2g07671
Length=255
Score = 35.8 bits (81), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 23/35 (65%), Gaps = 0/35 (0%)
Query 128 GIGSLFAALVSGTARNPSIKEDLFTYTLIGMGFLE 162
GIG++F++L+ ARNPS+ + F Y ++G E
Sbjct 34 GIGNVFSSLIHSVARNPSLAKQSFGYAILGFALTE 68
> At1g60070
Length=867
Score = 32.0 bits (71), Expect = 0.65, Method: Composition-based stats.
Identities = 26/90 (28%), Positives = 37/90 (41%), Gaps = 15/90 (16%)
Query 28 AAAALAAPKAAPSFPFCSSFC--------GSFCPTLAFRALSTAPLLQRHSAVAQGPCAR 79
A A +A K + FP CS GSF L R+L + ++Q+H + R
Sbjct 535 AMALIALLKISSRFPSCSERVKSIIGQNKGSFVLELQQRSLEFSSVIQKHQNIRSSLVER 594
Query 80 WPAAALTPLSS-------SSSSSSGGSPVG 102
P S +S S+SG SP+G
Sbjct 595 MPVLDEATFSGRRAGSLPASVSTSGKSPLG 624
> CE23569
Length=1809
Score = 31.6 bits (70), Expect = 0.85, Method: Composition-based stats.
Identities = 35/104 (33%), Positives = 48/104 (46%), Gaps = 21/104 (20%)
Query 45 SSFCGS-FCPTLAFRALSTAPLLQRHSAVA-------QGPCARWPAAALTPL--SSSSSS 94
+SFC + L RA L+Q AVA +GP ARW + L PL S++
Sbjct 623 TSFCNAPLNGLLTRRAREAGGLIQ---AVAPWFHPSMEGPAARW-SEKLEPLLDELSTTV 678
Query 95 SSGGSPVGAVR-------HEASVATLSAAVALMSVGGVAQGIGS 131
SSG S +R HEA + LSA VA++ G Q + +
Sbjct 679 SSGDSAPAELRGRKIARWHEACLDWLSACVAVVPEGDWRQDLAA 722
> Hs10198656
Length=573
Score = 30.4 bits (67), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 33/70 (47%), Gaps = 7/70 (10%)
Query 24 CSRAAAAALAAPKAAPSFPF-CSSFCGSFCPT--LAFRALSTAPLL-QR---HSAVAQGP 76
C + AALA P+ + FP C C CP + A + P QR H +V QGP
Sbjct 500 CVQQRLAALAHPQESHPFPESCPRGCPPLCPEDCTSIPAPTILPCRPQRSACHFSVQQGP 559
Query 77 CARWPAAALT 86
C+R P A T
Sbjct 560 CSRNPQPACT 569
> SPCC4B3.10c
Length=640
Score = 29.3 bits (64), Expect = 4.5, Method: Composition-based stats.
Identities = 23/83 (27%), Positives = 39/83 (46%), Gaps = 5/83 (6%)
Query 69 HSAVAQGPCARWPAAALTPLSSSSSSSSGGSPVGAVRHEASVATLSAAVALMSVGGVAQG 128
+S +AQ +L+P++S++SSS SPV H S +S S+ ++ G
Sbjct 286 NSLIAQNLHTSASQVSLSPMASTASSSVTNSPVDT--HTPSTPIMSRPP---SMKALSSG 340
Query 129 IGSLFAALVSGTARNPSIKEDLF 151
+ S ++ S + P I LF
Sbjct 341 VESQDESVASSNFQVPIISNPLF 363
> AtMi003
Length=315
Score = 28.5 bits (62), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 10/19 (52%), Positives = 16/19 (84%), Gaps = 0/19 (0%)
Query 128 GIGSLFAALVSGTARNPSI 146
GIG++F++L+ ARNPS+
Sbjct 34 GIGNVFSSLIHSVARNPSL 52
> SPAC4G9.04c
Length=638
Score = 28.5 bits (62), Expect = 7.2, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 26/61 (42%), Gaps = 0/61 (0%)
Query 35 PKAAPSFPFCSSFCGSFCPTLAFRALSTAPLLQRHSAVAQGPCARWPAAALTPLSSSSSS 94
P P+ P S S P L + P ++S + P + + A ALTP SSS
Sbjct 301 PSTIPTIPSAYSASVSSQPPLTHSYVHPGPQSHKYSLSSGPPASLYNANALTPEESSSID 360
Query 95 S 95
S
Sbjct 361 S 361
> YOL129w
Length=184
Score = 28.5 bits (62), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 148 EDLFTYTLIGMGFLEFLGIICVMMSAVLLY 177
+D TY +GMG LG +C+++S V+L+
Sbjct 140 KDYNTYPTLGMGVNNVLGNVCILLSCVVLW 169
> AtCh010
Length=81
Score = 28.5 bits (62), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 38/72 (52%), Gaps = 3/72 (4%)
Query 108 ASVATLSAAVALMSVG-GVAQGIGSLFAALVSGTARNPSIKEDLFTYTLIGMGFLEFLGI 166
ASV AV L S+G GV QG + A V G AR P + + L+ + F+E L I
Sbjct 8 ASVIAAGLAVGLASIGPGVGQGTAAGQA--VEGIARQPEAEGKIRGTLLLSLAFMEALTI 65
Query 167 ICVMMSAVLLYS 178
++++ LL++
Sbjct 66 YGLVVALALLFA 77
Lambda K H
0.321 0.130 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2779358582
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40