bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2777_orf1
Length=162
Score E
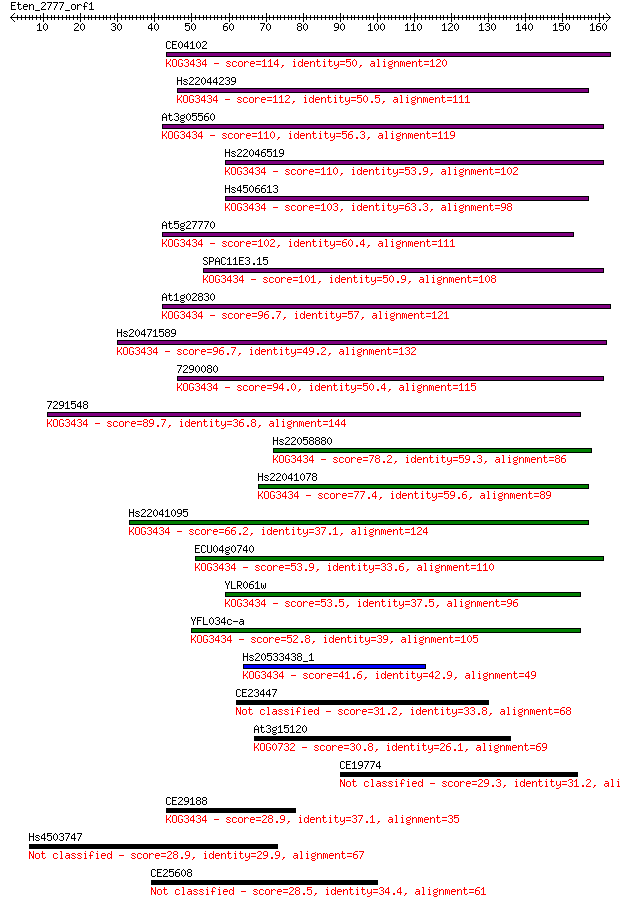
Sequences producing significant alignments: (Bits) Value
CE04102 114 9e-26
Hs22044239 112 2e-25
At3g05560 110 1e-24
Hs22046519 110 2e-24
Hs4506613 103 2e-22
At5g27770 102 4e-22
SPAC11E3.15 101 6e-22
At1g02830 96.7 2e-20
Hs20471589 96.7 2e-20
7290080 94.0 1e-19
7291548 89.7 2e-18
Hs22058880 78.2 6e-15
Hs22041078 77.4 1e-14
Hs22041095 66.2 3e-11
ECU04g0740 53.9 1e-07
YLR061w 53.5 2e-07
YFL034c-a 52.8 3e-07
Hs20533438_1 41.6 6e-04
CE23447 31.2 0.89
At3g15120 30.8 1.4
CE19774 29.3 3.4
CE29188 28.9 4.1
Hs4503747 28.9 4.3
CE25608 28.5 6.3
> CE04102
Length=130
Score = 114 bits (285), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 60/121 (49%), Positives = 80/121 (66%), Gaps = 2/121 (1%)
Query 43 SRGKKAATAGKATKIKFVIDCQKPVEDNILEAKGLEKFLQSRIKVAGKLNNFG-EKIEVF 101
++ K A K +KF ++C+ PVED IL + LE FL +IKV GK + ++V
Sbjct 7 AKSAKKALRKKKVHLKFNVECKNPVEDGILRIEDLEAFLNEKIKVNGKTGHLAANNVKVE 66
Query 102 REKAKVCVTAELPFSKRYLKYLVKKYLKKQMLRDFLRVVANNKTSYELRYFQMPQDDEEA 161
K+KV V +E+PFSKRYLKYL KKYLK+ LRD+LRVVA NK +YE+RYF + D E+A
Sbjct 67 VAKSKVSVVSEVPFSKRYLKYLTKKYLKRNSLRDWLRVVAVNKNTYEVRYFHI-NDGEDA 125
Query 162 A 162
Sbjct 126 G 126
> Hs22044239
Length=141
Score = 112 bits (281), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 56/114 (49%), Positives = 78/114 (68%), Gaps = 3/114 (2%)
Query 46 KKAATAGKATK--IKFVIDCQKPVEDNILEAKGLEKFLQSRIKVAGKLNNFGEKI-EVFR 102
K G+ TK ++F + C P+ED I++A E+FLQ RIKV GK+ N G + + +
Sbjct 19 KACGEGGQKTKQVLRFTLGCTHPIEDGIIDAANFEQFLQERIKVNGKVGNLGGGVVTIEK 78
Query 103 EKAKVCVTAELPFSKRYLKYLVKKYLKKQMLRDFLRVVANNKTSYELRYFQMPQ 156
K+K+ VT+++PFSKRYL YL KKYLKK L D+L +VAN+K SY+L YFQ+ Q
Sbjct 79 SKSKITVTSKVPFSKRYLIYLTKKYLKKNNLHDWLCIVANSKESYKLCYFQINQ 132
> At3g05560
Length=124
Score = 110 bits (275), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 67/121 (55%), Positives = 83/121 (68%), Gaps = 2/121 (1%)
Query 42 MSRGKKAATAGKATKIKFVIDCQKPVEDNILEAKGLEKFLQSRIKVAGKLNNFGEKIEVF 101
MSRG A GK + F IDC KPV+D I+E LEKFLQ RIKV GK G+ + +
Sbjct 1 MSRGGAAVAKGKKKGVSFTIDCSKPVDDKIMEIASLEKFLQERIKVGGKAGALGDSVTIT 60
Query 102 REKAKVCVTAELPFSKRYLKYLVKKYLKKQMLRDFLRVVANNKTS--YELRYFQMPQDDE 159
REK+K+ VTA+ FSKRYLKYL KKYLKK +RD+LRV+A NK YELRYF + +++
Sbjct 61 REKSKITVTADGQFSKRYLKYLTKKYLKKHNVRDWLRVIAANKDRNLYELRYFNIAENEG 120
Query 160 E 160
E
Sbjct 121 E 121
> Hs22046519
Length=126
Score = 110 bits (274), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 55/103 (53%), Positives = 71/103 (68%), Gaps = 2/103 (1%)
Query 59 FVIDCQKPVEDNILEAKGLEKFLQSRIKVAGKLNNFGEKIEVF-REKAKVCVTAELPFSK 117
F ++C PVED I++A E+FLQ RIKV GK N G + R K+K+ VT+E+PFSK
Sbjct 21 FTLECTHPVEDGIMDAANFEQFLQERIKVNGKAENLGGGMATIERSKSKINVTSEVPFSK 80
Query 118 RYLKYLVKKYLKKQMLRDFLRVVANNKTSYELRYFQMPQDDEE 160
RY KYL K Y+K LRD+L +V N+K SY+L YFQ+ QD EE
Sbjct 81 RYFKYLTKTYMKNN-LRDWLLLVTNSKDSYKLCYFQINQDKEE 122
> Hs4506613
Length=128
Score = 103 bits (257), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 62/99 (62%), Positives = 75/99 (75%), Gaps = 1/99 (1%)
Query 59 FVIDCQKPVEDNILEAKGLEKFLQSRIKVAGKLNNFGEKI-EVFREKAKVCVTAELPFSK 117
F +DC PVED I++A E+FLQ RIKV GK N G + + R K+K+ VT+E+PFSK
Sbjct 21 FTLDCTHPVEDGIMDAANFEQFLQERIKVNGKAGNLGGGVVTIERSKSKITVTSEVPFSK 80
Query 118 RYLKYLVKKYLKKQMLRDFLRVVANNKTSYELRYFQMPQ 156
RYLKYL KKYLKK LRD+LRVVAN+K SYELRYFQ+ Q
Sbjct 81 RYLKYLTKKYLKKNNLRDWLRVVANSKESYELRYFQINQ 119
> At5g27770
Length=124
Score = 102 bits (253), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 67/113 (59%), Positives = 79/113 (69%), Gaps = 2/113 (1%)
Query 42 MSRGKKAATAGKATKIKFVIDCQKPVEDNILEAKGLEKFLQSRIKVAGKLNNFGEKIEVF 101
MSRG AA GK + F IDC KPV+D I+E LEKFLQ RIKV GK G+ + +
Sbjct 1 MSRGNAAAAKGKKKGVSFTIDCSKPVDDKIMEIASLEKFLQERIKVGGKAGALGDSVSIT 60
Query 102 REKAKVCVTAELPFSKRYLKYLVKKYLKKQMLRDFLRVVANNKTS--YELRYF 152
REK+K+ VTA+ FSKRYLKYL KKYLKK +RD+LRV+A NK YELRYF
Sbjct 61 REKSKITVTADGQFSKRYLKYLTKKYLKKHNVRDWLRVIAANKDRNLYELRYF 113
> SPAC11E3.15
Length=117
Score = 101 bits (251), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 55/113 (48%), Positives = 70/113 (61%), Gaps = 5/113 (4%)
Query 53 KATKI--KFVIDCQKPVEDNILEAKGLEKFLQSRIKVAGKLNNFGEKIEVFRE-KAKVCV 109
K TK+ K++ID V D I + EK+L RIKV GK N G + V RE +K+ V
Sbjct 4 KNTKVSNKYIIDATAAVNDKIFDVAAFEKYLIDRIKVDGKTGNLGSSVVVSREGSSKIAV 63
Query 110 TAELPFSKRYLKYLVKKYLKKQMLRDFLRVVANNKTSYELRYFQ--MPQDDEE 160
A + FS RYLKYL KK+LKK LRD+LRVV+ K YELRY+ + D+EE
Sbjct 64 IAHIDFSGRYLKYLTKKFLKKHSLRDWLRVVSTKKGVYELRYYNVVVGNDEEE 116
> At1g02830
Length=127
Score = 96.7 bits (239), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 69/125 (55%), Positives = 84/125 (67%), Gaps = 6/125 (4%)
Query 42 MSR-GKKAATAGKATK-IKFVIDCQKPVEDNILEAKGLEKFLQSRIKVAGKLNNFGEKIE 99
M+R G K++ AG K + FVIDC KPV+D ILE LEKFLQ RIKV GK G +
Sbjct 1 MARVGAKSSGAGAKKKGVSFVIDCSKPVDDTILEIATLEKFLQERIKVRGKAGALGNSVS 60
Query 100 VFREKAKVCVTAELPFSKRYLKYLVKKYLKKQMLRDFLRVVANN--KTSYELRYFQMPQD 157
+ R K+ V A FSKRYLKYL KKYLKK LRD+LRV+A+N K YE+RYF++ D
Sbjct 61 ITRYNGKINVNANSNFSKRYLKYLTKKYLKKYNLRDWLRVIASNKDKNVYEVRYFRI--D 118
Query 158 DEEAA 162
DE A+
Sbjct 119 DEVAS 123
> Hs20471589
Length=182
Score = 96.7 bits (239), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 65/137 (47%), Positives = 88/137 (64%), Gaps = 5/137 (3%)
Query 30 VSASGAAQARSHMSRGKKAATAGKATKIK-----FVIDCQKPVEDNILEAKGLEKFLQSR 84
++A AA A S +SR + K K K F +D PVED I ++ E+FL+ +
Sbjct 42 INADRAAVASSLLSRPTRKMAPQKDRKPKRSTWRFNLDLTHPVEDGIFDSGNFEQFLREK 101
Query 85 IKVAGKLNNFGEKIEVFREKAKVCVTAELPFSKRYLKYLVKKYLKKQMLRDFLRVVANNK 144
+KV GK N G + + R K K+ V +E FSKRYLKYL KKYLKK LRD+LRVVA++K
Sbjct 102 VKVNGKTGNLGNVVHIERFKNKITVVSEKQFSKRYLKYLTKKYLKKNNLRDWLRVVASDK 161
Query 145 TSYELRYFQMPQDDEEA 161
+YELRYFQ+ QD++E+
Sbjct 162 ETYELRYFQISQDEDES 178
> 7290080
Length=299
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 58/120 (48%), Positives = 81/120 (67%), Gaps = 5/120 (4%)
Query 46 KKAATAGKATK-----IKFVIDCQKPVEDNILEAKGLEKFLQSRIKVAGKLNNFGEKIEV 100
KK GK K ++F IDC ED+I++ EK++++R+KV GK+NN G +
Sbjct 175 KKNVLRGKGQKKKKVSLRFTIDCTNIAEDSIMDVADFEKYIKARLKVNGKVNNLGNNVTF 234
Query 101 FREKAKVCVTAELPFSKRYLKYLVKKYLKKQMLRDFLRVVANNKTSYELRYFQMPQDDEE 160
R K K+ V++++ FSK YLKYL KKYLKK LRD++RVVAN K SYELRYF++ +D+E
Sbjct 235 ERSKLKLIVSSDVHFSKAYLKYLTKKYLKKNSLRDWIRVVANEKDSYELRYFRISSNDDE 294
> 7291548
Length=312
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 53/145 (36%), Positives = 83/145 (57%), Gaps = 3/145 (2%)
Query 11 SVDVPFVGASSTDFCVATMVSASGAAQARSHMSRGKKAATAGKATKIKFVIDCQKPVEDN 70
+V+VP A S +AT + + ++ RGK+ A K +FVIDC ED
Sbjct 148 NVEVPAAKAKSVKKSLATPANPKPKPKRTKNVLRGKR--LAKKKAWQRFVIDCACVAEDM 205
Query 71 ILEAKGLEKFLQSRIKVAGKLNNFGEKIEVFREKA-KVCVTAELPFSKRYLKYLVKKYLK 129
IL+ E++L++ IK+ KLN +++ R K + + + + FSKRY KYL K+YLK
Sbjct 206 ILDLADFEQYLKTHIKIKNKLNQLKDQVTFERTKNFSLIIHSGVHFSKRYFKYLTKRYLK 265
Query 130 KQMLRDFLRVVANNKTSYELRYFQM 154
K LRD+LRVV+ K ++ + YF++
Sbjct 266 KVSLRDWLRVVSTAKDTFAMTYFKI 290
> Hs22058880
Length=94
Score = 78.2 bits (191), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 51/87 (58%), Positives = 62/87 (71%), Gaps = 1/87 (1%)
Query 72 LEAKGLEKFLQSRIKVAGKLNNFGEKIEVFR-EKAKVCVTAELPFSKRYLKYLVKKYLKK 130
++A E+F Q RIK+ GK NFG + K+K VT++LPFS RYLKYL KKYLKK
Sbjct 1 MDAANFEQFFQERIKMNGKAGNFGGGVVTIEGSKSKTSVTSKLPFSNRYLKYLTKKYLKK 60
Query 131 QMLRDFLRVVANNKTSYELRYFQMPQD 157
L D+LRVVAN+K SYELRYFQ+ QD
Sbjct 61 NNLHDWLRVVANSKQSYELRYFQINQD 87
> Hs22041078
Length=150
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 53/90 (58%), Positives = 67/90 (74%), Gaps = 1/90 (1%)
Query 68 EDNILEAKGLEKFLQSRIKVAGKLNNFGEK-IEVFREKAKVCVTAELPFSKRYLKYLVKK 126
ED I++A E+FLQ RIKV GK N GE + V R K+K+ VT+++ FSKRYLKYL KK
Sbjct 52 EDGIVDAANFEQFLQERIKVNGKAGNLGEGVVTVKRSKSKITVTSKVHFSKRYLKYLTKK 111
Query 127 YLKKQMLRDFLRVVANNKTSYELRYFQMPQ 156
YLKK LRD+L +VAN+K SY+LR FQ+ Q
Sbjct 112 YLKKNNLRDWLCIVANSKESYKLRSFQINQ 141
> Hs22041095
Length=326
Score = 66.2 bits (160), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 46/130 (35%), Positives = 64/130 (49%), Gaps = 31/130 (23%)
Query 33 SGAAQARSHMSRGK--KAATAGKATK---IKFVIDCQKPVEDNILEAKGLEKFLQSRIKV 87
S A + R SR + KA G K ++F + C P+ED I++A E+FLQ RIKV
Sbjct 213 SNAVEGRKDFSRAQIQKACGEGGQKKKQVLRFTLGCTHPIEDGIVDAANFEQFLQERIKV 272
Query 88 AGKLNNFGEKI-EVFREKAKVCVTAELPFSKRYLKYLVKKYLKKQMLRDFLRVVANNKTS 146
GK N GE + V R K+K+ VT+++ FSK +K S
Sbjct 273 NGKAGNLGEGVVTVKRSKSKITVTSKVHFSK-------------------------SKES 307
Query 147 YELRYFQMPQ 156
Y+LR FQ+ Q
Sbjct 308 YKLRSFQINQ 317
> ECU04g0740
Length=112
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 37/117 (31%), Positives = 57/117 (48%), Gaps = 15/117 (12%)
Query 51 AGKATKIKFVIDCQKPVEDNILEAKGLEKFLQSRIKV-AGK------LNNFGEKIEVFRE 103
A + T +F IDC KP D+++ L FLQ +IK GK +N G +E
Sbjct 4 ASEKTTRRFTIDCTKPASDSLISPSDLGAFLQQKIKCYTGKKEKLLHINANGNIVE---- 59
Query 104 KAKVCVTAELPFSKRYLKYLVKKYLKKQMLRDFLRVVANNKTSYELRYFQMPQDDEE 160
V VT K+ LK+ + ++L + LR F+++ A +ELRY + + EE
Sbjct 60 ---VDVTGGF-IGKQGLKWQIGRFLHMKKLRAFIKIFAQGLDGFELRYINVEEGKEE 112
> YLR061w
Length=121
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 36/96 (37%), Positives = 52/96 (54%), Gaps = 0/96 (0%)
Query 59 FVIDCQKPVEDNILEAKGLEKFLQSRIKVAGKLNNFGEKIEVFREKAKVCVTAELPFSKR 118
F +D P E+ + + K+L IKV G + N G + V + V V + FS +
Sbjct 15 FTVDVSSPTENGVFDPASYAKYLIDHIKVEGAVGNLGNAVTVTEDGTVVTVVSTAKFSGK 74
Query 119 YLKYLVKKYLKKQMLRDFLRVVANNKTSYELRYFQM 154
YLKYL KKYLKK LRD++R V+ Y L ++Q+
Sbjct 75 YLKYLTKKYLKKNQLRDWIRFVSTKTNEYRLAFYQV 110
> YFL034c-a
Length=122
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 41/106 (38%), Positives = 60/106 (56%), Gaps = 1/106 (0%)
Query 50 TAGKATKIK-FVIDCQKPVEDNILEAKGLEKFLQSRIKVAGKLNNFGEKIEVFREKAKVC 108
T+ K IK +D P E+ + + K+L IKV G + N G IEV + + V
Sbjct 5 TSRKQKVIKTLTVDVSSPTENGVFDPASYSKYLIDHIKVDGAVGNLGNAIEVTEDGSIVT 64
Query 109 VTAELPFSKRYLKYLVKKYLKKQMLRDFLRVVANNKTSYELRYFQM 154
V + FS +YLKYL KKYLKK LRD++R V+ + Y+L ++Q+
Sbjct 65 VVSSAKFSGKYLKYLTKKYLKKNQLRDWIRFVSIRQNQYKLVFYQV 110
> Hs20533438_1
Length=65
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 21/50 (42%), Positives = 33/50 (66%), Gaps = 1/50 (2%)
Query 64 QKPVEDNILEAKGLEKFLQSRIKVAGKLNNFGEK-IEVFREKAKVCVTAE 112
+K ED I++A E+FLQ RIKV GK+ N G + + + K+K+ VT++
Sbjct 13 KKKAEDGIVDAANFEQFLQERIKVNGKVGNLGGGVVTIEKSKSKITVTSK 62
> CE23447
Length=553
Score = 31.2 bits (69), Expect = 0.89, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 33/73 (45%), Gaps = 6/73 (8%)
Query 62 DCQKPVEDNILEAKGLEKF---LQSRIKVAGKLNNFGEKIEVFREKAKVCV--TAELPFS 116
DC K + + I +E F Q +K K+NN + + F K C AE +S
Sbjct 232 DCTKSIMEGICGKSAVENFDQYAQISVKNLNKMNNLAAQKKSFI-NGKACFFNIAEYNYS 290
Query 117 KRYLKYLVKKYLK 129
++KYL K Y K
Sbjct 291 DEHIKYLKKNYDK 303
> At3g15120
Length=1954
Score = 30.8 bits (68), Expect = 1.4, Method: Composition-based stats.
Identities = 18/69 (26%), Positives = 36/69 (52%), Gaps = 1/69 (1%)
Query 67 VEDNILEAKGLEKFLQSRIKVAGKLNNFGEKIEVFREKAKVCVTAELPFSKRYLKYLVKK 126
V++N E G EK LQ+ ++V+ N F E++E R K+ + A + L +++
Sbjct 1232 VQENCSEM-GEEKALQNGVRVSHAWNTFFEQVETLRVSTKMMILATSGMPYKLLPPKIQQ 1290
Query 127 YLKKQMLRD 135
+ K + ++
Sbjct 1291 FFKTDLSKE 1299
> CE19774
Length=785
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 28/64 (43%), Gaps = 0/64 (0%)
Query 90 KLNNFGEKIEVFREKAKVCVTAELPFSKRYLKYLVKKYLKKQMLRDFLRVVANNKTSYEL 149
KLN FG+ I+ E K V+ P K+ K Y+ K+ + L VV N Y +
Sbjct 438 KLNYFGDIIKWCNETRKSSVSINFPSPKQLASLHEKSYVLKKHMDSVLIVVNNYPWKYGM 497
Query 150 RYFQ 153
Q
Sbjct 498 GLIQ 501
> CE29188
Length=53
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 43 SRGKKAATAGKATKIKFVIDCQKPVEDNILEAKGL 77
++ K A K +KF ++C+ PVED IL + L
Sbjct 7 AKSAKKALRKKKVHLKFNVECKNPVEDGILRIEDL 41
> Hs4503747
Length=2602
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 31/67 (46%), Gaps = 2/67 (2%)
Query 6 GNKLLSVDVPFVGASSTDFCVATMVSASGAAQARSHMSRGKKAATAGKATKIKFVIDCQK 65
N+ S+ V V SST+ C + + AS A + G A G+ K F++DC K
Sbjct 2482 ANETSSILVESVTRSSTETCYSAIPKASSDASKVTSKGAGLSKAFVGQ--KSSFLVDCSK 2539
Query 66 PVEDNIL 72
+ +L
Sbjct 2540 AGSNMLL 2546
> CE25608
Length=580
Score = 28.5 bits (62), Expect = 6.3, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 32/65 (49%), Gaps = 4/65 (6%)
Query 39 RSHMSRGKKAAT--AGKATKIKFVIDCQKPVEDNILEA--KGLEKFLQSRIKVAGKLNNF 94
R MS G ++ T K ++F ++ K E E+ K KF + R+K K NF
Sbjct 505 RKRMSTGARSLTFCEIKPKNLRFWLELLKKKESRNSESFQKNWAKFFEFRLKFKIKNKNF 564
Query 95 GEKIE 99
G+K+E
Sbjct 565 GKKVE 569
Lambda K H
0.318 0.132 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2244926132
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40