bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2779_orf1
Length=215
Score E
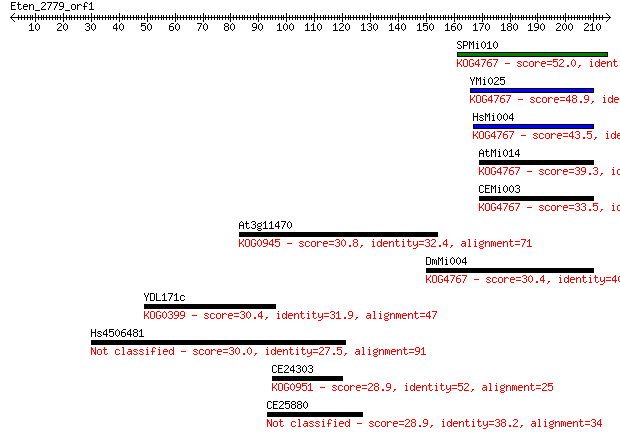
Sequences producing significant alignments: (Bits) Value
SPMi010 52.0 9e-07
YMi025 48.9 7e-06
HsMi004 43.5 3e-04
AtMi014 39.3 0.006
CEMi003 33.5 0.35
At3g11470 30.8 2.1
DmMi004 30.4 2.6
YDL171c 30.4 2.8
Hs4506481 30.0 3.8
CE24303 28.9 7.1
CE25880 28.9 7.6
> SPMi010
Length=248
Score = 52.0 bits (123), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 34/54 (62%), Gaps = 0/54 (0%)
Query 161 AEFLFFWTPTFIIWSLTIPVFTMLYMVDEAVYTAMTVKVIGRQWYWIYEVESPV 214
EF++ P I+ + +P F +LY++DE +MTVK IGRQW+W YE+ V
Sbjct 77 VEFIWTLIPALILILVALPSFKLLYLLDEVQKPSMTVKAIGRQWFWSYELNDFV 130
> YMi025
Length=251
Score = 48.9 bits (115), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 22/46 (47%), Positives = 28/46 (60%), Gaps = 2/46 (4%)
Query 166 FWT--PTFIIWSLTIPVFTMLYMVDEAVYTAMTVKVIGRQWYWIYE 209
WT P I+ + P F +LY+ DE + AMT+K IG QWYW YE
Sbjct 84 IWTIFPAVILLIIAFPSFILLYLCDEVISPAMTIKAIGYQWYWKYE 129
> HsMi004
Length=227
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 20/45 (44%), Positives = 27/45 (60%), Gaps = 2/45 (4%)
Query 167 WT--PTFIIWSLTIPVFTMLYMVDEAVYTAMTVKVIGRQWYWIYE 209
WT P I+ + +P +LYM DE ++T+K IG QWYW YE
Sbjct 65 WTILPAIILVLIALPSLRILYMTDEVNDPSLTIKSIGHQWYWTYE 109
> AtMi014
Length=260
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 20/42 (47%), Positives = 26/42 (61%), Gaps = 1/42 (2%)
Query 169 PTFIIWSLTIPVFTMLYMVDEAVYT-AMTVKVIGRQWYWIYE 209
P+ I + IP F +LY +DE V A+T+K IG QWY YE
Sbjct 89 PSIISMFIAIPSFALLYSMDEVVVDPAITIKAIGHQWYRTYE 130
> CEMi003
Length=231
Score = 33.5 bits (75), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 24/42 (57%), Gaps = 1/42 (2%)
Query 169 PTFIIWSLTIPVFTMLYMVDEA-VYTAMTVKVIGRQWYWIYE 209
PT I+ +P ++LY + + +TVKV G QWYW YE
Sbjct 71 PTIILLMQMVPSLSLLYYYGLMNLDSNLTVKVTGHQWYWSYE 112
> At3g11470
Length=275
Score = 30.8 bits (68), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 39/76 (51%), Gaps = 8/76 (10%)
Query 83 FINPDGTYTYPITSHLFHFEDTYSPVPKQQIVSINGKKMIK-GIESRSLVE-LFSIHQLN 140
FI PD + TS L H+ SP K++++ + G ++ K + +R+LV + +Q N
Sbjct 30 FIRPDEVKS---TSLLKHYSQLLSPTEKEKVLQMRGDELKKNALLARTLVRTTIARYQTN 86
Query 141 IPFFPRKRM---NVYG 153
P+ M N+YG
Sbjct 87 NEVDPKALMFKKNMYG 102
> DmMi004
Length=228
Score = 30.4 bits (67), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 24/62 (38%), Positives = 33/62 (53%), Gaps = 2/62 (3%)
Query 150 NVYGNHDLLMKAEFLFFWT--PTFIIWSLTIPVFTMLYMVDEAVYTAMTVKVIGRQWYWI 207
N Y N LL WT P I+ + +P +LY++DE ++T+K IG QWYW
Sbjct 48 NNYVNRFLLHGQLIEMIWTILPAIILLFIALPSLRLLYLLDEINEPSVTLKSIGHQWYWS 107
Query 208 YE 209
YE
Sbjct 108 YE 109
> YDL171c
Length=2145
Score = 30.4 bits (67), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 23/47 (48%), Gaps = 0/47 (0%)
Query 49 PGSAQKHEPQQHLEEPKGLYNMPQDHSHHGDPRKFINPDGTYTYPIT 95
P S Q + + +E+ +G + H H DPR +N +T PIT
Sbjct 1626 PDSKQLEKNSERIEKTRGFMIHKRRHETHRDPRTRVNDWKEFTNPIT 1672
> Hs4506481
Length=1224
Score = 30.0 bits (66), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 25/95 (26%), Positives = 40/95 (42%), Gaps = 13/95 (13%)
Query 30 PGAACARLGFST---NAPAQHSPGSAQKHEPQ-QHLEEPKGLYNMPQDHSHHGDPRKFIN 85
P C +L T A + P S H + EEP+ + P H +H +PR +
Sbjct 941 PSKDCPKLDMLTRNFQAATESVPNSGNIHNGSLEQTEEPETSSHSPSRHMNHSEPRPGLG 1000
Query 86 PDGTYTYPITSHLFHFEDTYSPVPKQQIVSINGKK 120
DG P+ + D+ +PK+ +NGK+
Sbjct 1001 ADGDAADPVDTR-----DSKFLLPKE----VNGKQ 1026
> CE24303
Length=1002
Score = 28.9 bits (63), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 18/26 (69%), Gaps = 1/26 (3%)
Query 95 TSHLFHFEDTYSPVP-KQQIVSINGK 119
T HL F+++Y PVP KQQI+ + K
Sbjct 440 TEHLHFFDNSYRPVPLKQQIIGVTEK 465
> CE25880
Length=354
Score = 28.9 bits (63), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 17/34 (50%), Gaps = 0/34 (0%)
Query 93 PITSHLFHFEDTYSPVPKQQIVSINGKKMIKGIE 126
P+ HL +FED Y P + +NGK I I
Sbjct 176 PLAQHLVNFEDIYCIGPFYYLKDVNGKSYINWIS 209
Lambda K H
0.323 0.138 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3969005466
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40