bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2789_orf3
Length=336
Score E
Sequences producing significant alignments: (Bits) Value
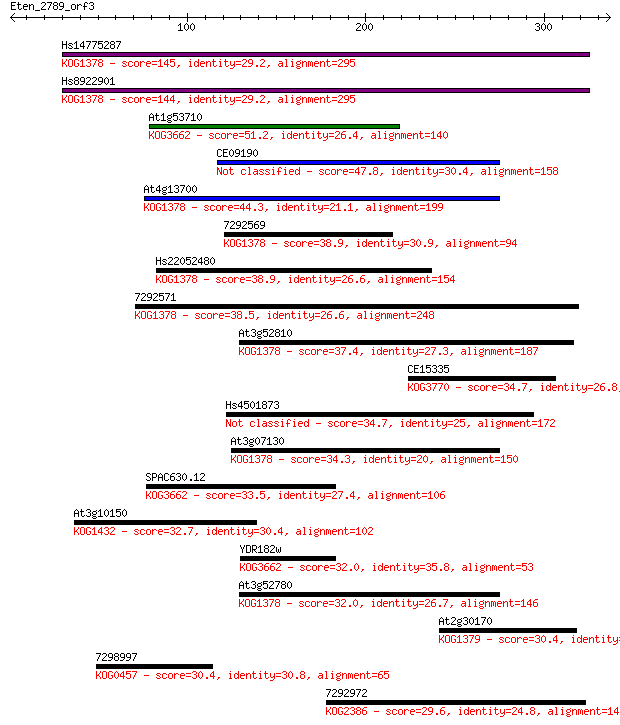
Hs14775287 145 1e-34
Hs8922901 144 3e-34
At1g53710 51.2 3e-06
CE09190 47.8 3e-05
At4g13700 44.3 4e-04
7292569 38.9 0.015
Hs22052480 38.9 0.017
7292571 38.5 0.022
At3g52810 37.4 0.049
CE15335 34.7 0.28
Hs4501873 34.7 0.32
At3g07130 34.3 0.41
SPAC630.12 33.5 0.69
At3g10150 32.7 1.3
YDR182w 32.0 1.8
At3g52780 32.0 2.0
At2g30170 30.4 5.5
7298997 30.4 5.8
7292972 29.6 8.7
> Hs14775287
Length=314
Score = 145 bits (366), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 86/304 (28%), Positives = 141/304 (46%), Gaps = 36/304 (11%)
Query 30 AEDQLTWSSPFTFALIADPQFGLYR---------NNNSWEEERQQVAECIIAAASLSSKP 80
AE + W PF F L ADPQFGL + + WE+E + + + A L+ KP
Sbjct 23 AEKESEWKGPFYFILGADPQFGLIKAWSTGDCDNGGDEWEQEIRLTEQAVQAINKLNPKP 82
Query 81 AFIAILGDLVHAPVPALSQESLQDEVRAVRDQQAKDLKDVIDKPSKDVPVLVLPGNHDVG 140
F + GDL+HA +P + R +Q +DLK V+ + +P++++ GNHD+G
Sbjct 83 KFFVLCGDLIHA-MPG----------KPWRTEQTEDLKRVLRAVDRAIPLVLVSGNHDIG 131
Query 141 NAPTAASIADYEQLWGKDHFSFWFGGVKFVAANSSVLYDDTNAKELAEEQFEWIENELKD 200
N PTA ++ ++ + WG D+FSFW GGV F+ NS + + L + Q +W++ +L
Sbjct 132 NTPTAETVEEFCRTWGDDYFSFWVGGVLFLVLNSQFYENPSKCPSLKQAQDQWLDEQLSI 191
Query 201 GMEKGAKQQFLLQHHQFYWDNWGEADDKTNTVEVRQMRAHNPVSLFQLPKRWRERFQPLM 260
++ + + QH + ++ E DD F L K R++
Sbjct 192 ARQRHCQHAIVFQHIPLFLESIDEDDD----------------YYFNLSKSTRKKLADKF 235
Query 261 KKYNVAASFHGHLHYDSLLQDGDTPQIVTSSCGFATNDGRPGWRLVTVEESKFSHEFYPV 320
V F GH H ++ + +V+S+ G G R+V V K H +Y +
Sbjct 236 IHAGVKVVFSGHYHRNAGGTYQNLDMVVSSAIGCQLGRDPHGLRVVVVTAEKIVHRYYSL 295
Query 321 KWLT 324
L+
Sbjct 296 DELS 299
> Hs8922901
Length=314
Score = 144 bits (363), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 86/304 (28%), Positives = 140/304 (46%), Gaps = 36/304 (11%)
Query 30 AEDQLTWSSPFTFALIADPQFGLYR---------NNNSWEEERQQVAECIIAAASLSSKP 80
AE + W PF F L ADPQFGL + + WE+E + + + A L+ KP
Sbjct 23 AEKESEWKGPFYFILGADPQFGLIKAWSTGDCDNGGDEWEQEIRLTEQAVQAINELNPKP 82
Query 81 AFIAILGDLVHAPVPALSQESLQDEVRAVRDQQAKDLKDVIDKPSKDVPVLVLPGNHDVG 140
F + GDL+HA +P + R +Q +DLK V+ + +P++++ GNHD+G
Sbjct 83 KFFVLCGDLIHA-MPG----------KPWRTEQTEDLKRVLRAVDRAIPLVLVSGNHDIG 131
Query 141 NAPTAASIADYEQLWGKDHFSFWFGGVKFVAANSSVLYDDTNAKELAEEQFEWIENELKD 200
N PTA ++ ++ + WG D+FSFW GGV F+ NS + + L + Q +W++ +L
Sbjct 132 NTPTAETVEEFCRTWGDDYFSFWVGGVLFLVLNSQFYENPSKCPSLKQAQDQWLDEQLSI 191
Query 201 GMEKGAKQQFLLQHHQFYWDNWGEADDKTNTVEVRQMRAHNPVSLFQLPKRWRERFQPLM 260
++ + + QH + ++ E DD F L K R+
Sbjct 192 ARQRHCQHAIVFQHIPLFLESIDEDDD----------------YYFNLSKSTRKELADKF 235
Query 261 KKYNVAASFHGHLHYDSLLQDGDTPQIVTSSCGFATNDGRPGWRLVTVEESKFSHEFYPV 320
V F GH H ++ + +V+S+ G G R+V V K H +Y +
Sbjct 236 IHAGVRVVFSGHYHRNAGGTYQNLDMVVSSAIGCQLGRDPHGLRVVVVTAEKIVHRYYSL 295
Query 321 KWLT 324
L+
Sbjct 296 DELS 299
> At1g53710
Length=393
Score = 51.2 bits (121), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 37/140 (26%), Positives = 69/140 (49%), Gaps = 12/140 (8%)
Query 79 KPAFIAILGDLVHAPVPALSQESLQDEVRAVRDQQAKDLKDVIDKPSKDVPVLVLPGNHD 138
KP + LGD P LS+E Q+ + ++ + + + D+P +PGNHD
Sbjct 100 KPDVVLFLGDYFDGG-PFLSEEEWQESLNRLKHVFGLNSEGRVG----DIPTFYIPGNHD 154
Query 139 VGNAPTAASIADYEQLWGKDHFSFWFGGVKFVAANSSVLYDDTNAKELAEEQFEWIENEL 198
+G + I YE+++G + F G V+F++ ++ + D + K+LA E +++++N
Sbjct 155 IGYSRV---IDRYEKVFGVRNRRFMIGNVEFISIDAQAI-DGNSKKDLASEVWKFVQNVS 210
Query 199 KDGMEKGAKQQFLLQHHQFY 218
D + + LL H Y
Sbjct 211 TDAQ---SHPRVLLTHIPLY 227
> CE09190
Length=419
Score = 47.8 bits (112), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 48/190 (25%), Positives = 70/190 (36%), Gaps = 61/190 (32%)
Query 117 LKDVIDKPSKDVPVLVLPGNHD-VGNAPTAASIADYEQLWGKDHFS--FWFGGVKFVAAN 173
++V PS VP L + GNHD GN + + W +F ++ V+F +
Sbjct 143 FENVYTNPSLQVPWLTIAGNHDHFGNVTAEIEYTKHSKKW---YFPSLYYKKSVEFNGTS 199
Query 174 SSVLYDDT-----NAKEL------------------------AEEQFEWIENELKDGMEK 204
L DT N K++ AEEQ+ W+EN L E
Sbjct 200 IDFLMIDTISLCGNTKDIQNAGFIEMLRNESHDPRGPVNITAAEEQWAWLENNL----EA 255
Query 205 GAKQQFLLQHHQFYWDNWGEADDKTNTVEVRQMRAHNPVSLFQLPKRWRERFQPLMKKYN 264
+ Q ++ H V M +H P R+R PL+K++N
Sbjct 256 SSAQYLIISGH----------------YPVHSMSSHGPTDCL------RQRLDPLLKRFN 293
Query 265 VAASFHGHLH 274
V A F GH H
Sbjct 294 VNAYFSGHDH 303
> At4g13700
Length=474
Score = 44.3 bits (103), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 42/222 (18%), Positives = 89/222 (40%), Gaps = 44/222 (19%)
Query 76 LSSKPAFIAILGDLVHA---------PVPALSQESLQDEVRAVRDQQAKDLKDVIDKPSK 126
+ + P+ + I+GDL +A VP S +R + ++ +
Sbjct 215 MENDPSLVIIVGDLTYANQYRTIGGKGVPCFSCSFPDAPIRETYQPRWDAWGRFMEPLTS 274
Query 127 DVPVLVLPGNHDVGNAPTAASIADYEQLWG----------KDHFSFWFGGVKFVAANSSV 176
VP +V+ GNH++ + + Y + + ++SF GGV FV + V
Sbjct 275 KVPTMVIEGNHEIEPQASGITFKSYSERFAVPASESGSNSNLYYSFDAGGVHFVMLGAYV 334
Query 177 LYDDT----NAKELAEEQFEWIENELKDGMEKGAKQQFLLQHHQFYWDNWGEADDKTNTV 232
Y++T + E++ Q+ W++ +L +++ + H +++++
Sbjct 335 DYNNTGKSMDTLEVSWLQYAWLKEDLSK-VDRAVTPWLVATMHPPWYNSYS--------- 384
Query 233 EVRQMRAHNPVSLFQLPKRWRERFQPLMKKYNVAASFHGHLH 274
S +Q + R+ + L+ +Y V F GH+H
Sbjct 385 -----------SHYQEFECMRQEMEELLYQYRVDIVFAGHVH 415
> 7292569
Length=450
Score = 38.9 bits (89), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 29/101 (28%), Positives = 52/101 (51%), Gaps = 11/101 (10%)
Query 121 IDKPSKDVPVLVLPGNHD----VGNAPTAASI-ADYEQLWGKDHFSFWFGGVKFVAANSS 175
I+ + VP +V PGNH+ N ++ + + LW +SF G V FV+ ++
Sbjct 197 IESVAAYVPYMVCPGNHEEKYNFSNYRARFNMPGETDSLW----YSFNLGPVHFVSFSTE 252
Query 176 VLYDDTNAKELAEEQFEWIENELKDGM--EKGAKQQFLLQH 214
V Y + +L +QFEW+E +L + E AK+ +++ +
Sbjct 253 VYYFLSYGFKLLTKQFEWLERDLAEANLPENRAKRPWIITY 293
> Hs22052480
Length=588
Score = 38.9 bits (89), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 41/177 (23%), Positives = 75/177 (42%), Gaps = 27/177 (15%)
Query 83 IAILGDLVH---APVPALSQESLQDEVRAVR---------DQQAKDLKD----VIDKPSK 126
+A+ GDL VP L +++ Q AV DQ + D +I+ +
Sbjct 286 LAVFGDLGADNPKAVPRLRRDTQQGMYDAVLHVGDFAYNLDQDNARVGDRFMRLIEPVAA 345
Query 127 DVPVLVLPGNHD----VGNAPTAASI-ADYEQLWGKDHFSFWFGGVKFVAANSSVLYDDT 181
+P + PGNH+ N S+ D E LW +S+ G ++ ++ V +
Sbjct 346 SLPYMTCPGNHEERYNFSNYKARFSMPGDNEGLW----YSWDLGPAHIISFSTEVYFFLH 401
Query 182 NAKELAEEQFEWIENELKDGMEKGAKQQFLLQ--HHQFYWDNWGEADDKTNTVEVRQ 236
+ L + QF W+E++L+ + A + +++ H Y N D + +VR+
Sbjct 402 YGRHLVQRQFRWLESDLQKANKNRAARPWIITMGHRPMYCSNADLDDCTRHESKVRK 458
> 7292571
Length=448
Score = 38.5 bits (88), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 66/287 (22%), Positives = 112/287 (39%), Gaps = 61/287 (21%)
Query 71 IAAASLSSKPAFIAILGDLVHAPVPAL------SQESLQDEVRAVRD----------QQA 114
+ +AS+ P+ +AI GD+ + +L +Q + D + V D +
Sbjct 142 VPSASVDWSPS-LAIYGDMGNENAQSLARLQQETQRGMYDAIIHVGDFAYDMNTKNARVG 200
Query 115 KDLKDVIDKPSKDVPVLVLPGNHDVGNAPTAASIADYEQLWGKDHFSFWFGGVKFVAANS 174
+ I+ + +P +V+PGNH+ P E ++ +SF G V FV ++
Sbjct 201 DEFMRQIETVAAYLPYMVVPGNHEEKFMPGGT-----ENMF----YSFDLGPVHFVGIST 251
Query 175 SVLYDDTNAKELAEEQFEWIENEL-KDGMEKGAKQQ---FLLQHHQFYWDNWGEADDKTN 230
V Y + QFEW+ +L K + + ++ L H Y N E D+
Sbjct 252 EVYYFLNYGLKPLVFQFEWLREDLAKANLPENRNKRPWIILYGHRPMYCSN--ENDNDCT 309
Query 231 TVEVRQMRAHNPVSLFQLPKRWRERFQPLMKKYNVAASFHGHLH--------YDSLLQDG 282
E V +F L +PL+ ++ V + H H YD +++G
Sbjct 310 HSETLTRVGWPFVHMFGL--------EPLLYEFGVDVAIWAHEHSYERLWPIYDYKVRNG 361
Query 283 ---DTP--------QIVTSSCGFATNDGRPGWRLVTVEESKFSHEFY 318
D+P IVT S G +GR ++ E S F + Y
Sbjct 362 TLKDSPYNDPSAPVHIVTGSAG--CKEGREPFKGKIPEWSAFHSQDY 406
> At3g52810
Length=437
Score = 37.4 bits (85), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 51/202 (25%), Positives = 78/202 (38%), Gaps = 33/202 (16%)
Query 129 PVLVLPGNHDVGNAPTAASIA--DYEQLWGKDH----------FSFWFGGVKFVAANSSV 176
P +V GNH++ + PT I+ Y W H +SF GV V S
Sbjct 205 PWMVTEGNHEIESFPTNDHISFKSYNARWLMPHAESLSHSNLYYSFDVAGVHTVMLGSYT 264
Query 177 LYDDTNAKELAEEQFEWIENELKDGMEKGAKQQFLLQHHQFYWDN---WGEADDKTNTVE 233
Y E +Q+ W++ +L+ K ++ H +Y N +GE + + +E
Sbjct 265 PY------ESHSDQYHWLQADLRKVDRKKTPWLVVVMHTPWYSTNKAHYGEGEKMRSALE 318
Query 234 VRQMRAHNPVSLFQLPKRWRERFQPLMKKYNVAASFHGHLHYDSLLQDGDTPQIVTSSCG 293
RA V +F ERF+P+ YN A G ++ + DG + G
Sbjct 319 SLLYRAQVDV-VFAGHVHTYERFKPI---YNKKADPCGPMYI--TIGDGGNRE------G 366
Query 294 FATNDGRPGWRLVTVEESKFSH 315
A +P L ES F H
Sbjct 367 LALRFKKPQSPLSEFRESSFGH 388
> CE15335
Length=572
Score = 34.7 bits (78), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 22/84 (26%), Positives = 33/84 (39%), Gaps = 2/84 (2%)
Query 224 EADDKTNTVEVRQMRAHNPVSLFQLPKRWRERFQPLMKKY--NVAASFHGHLHYDSLLQD 281
E D N E+ + +H P K W F ++K+Y +A F+GH HYD +
Sbjct 411 ELQDSENKGELVHIISHIPPGDNYCLKGWSWNFFEIVKRYENTIAQMFYGHTHYDQFMTV 470
Query 282 GDTPQIVTSSCGFATNDGRPGWRL 305
D + + P W L
Sbjct 471 KDAKTYFANVTEANMKNKEPEWVL 494
> Hs4501873
Length=325
Score = 34.7 bits (78), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 43/192 (22%), Positives = 67/192 (34%), Gaps = 46/192 (23%)
Query 122 DKPSKDVPVLVLPGNHD-VGNAPTAASIADYEQLWG------KDHFS----------FWF 164
D+ + VP VL GNHD +GN + + + W + HF F
Sbjct 96 DRSLRKVPWYVLAGNHDHLGNVSAQIAYSKISKRWNFPSPFYRLHFKIPQTNVSVAIFML 155
Query 165 GGVKFVAANSSVLY---DDTNAKELAEEQFEWIENELKDGMEKGAKQQFLLQHHQFYWDN 221
V + L + +LA Q W++ +L A++ ++L ++
Sbjct 156 DTVTLCGNSDDFLSQQPERPRDVKLARTQLSWLKKQLA-----AAREDYVLV--AGHYPV 208
Query 222 WGEADDKTNTVEVRQMRAHNPVSLFQLPKRWRERFQPLMKKYNVAASFHGHLHYDSLLQD 281
W A+ V+Q+R PL+ Y V A GH H LQD
Sbjct 209 WSIAEHGPTHCLVKQLR-------------------PLLATYGVTAYLCGHDHNLQYLQD 249
Query 282 GDTPQIVTSSCG 293
+ V S G
Sbjct 250 ENGVGYVLSGAG 261
> At3g07130
Length=532
Score = 34.3 bits (77), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 30/160 (18%), Positives = 62/160 (38%), Gaps = 37/160 (23%)
Query 125 SKDVPVLVLPGNHDVGNAPTAASIADYEQLWG----------KDHFSFWFGGVKFVAANS 174
+ VP++V+ GNH++ + Y + ++SF GG+ FV +
Sbjct 266 TSKVPLMVIEGNHEIELQAENKTFEAYSSRFAFPFNESGSSSTLYYSFNAGGIHFVMLGA 325
Query 175 SVLYDDTNAKELAEEQFEWIENELKDGMEKGAKQQFLLQHHQFYWDNWGEADDKTNTVEV 234
+ YD + EQ+EW++ +L +++ + H ++ ++ T
Sbjct 326 YIAYDK------SAEQYEWLKKDLAK-VDRSVTPWLVASWHPPWYSSY--------TAHY 370
Query 235 RQMRAHNPVSLFQLPKRWRERFQPLMKKYNVAASFHGHLH 274
R+ +E + L+ Y F+GH+H
Sbjct 371 REAEC------------MKEAMEELLYSYGTDIVFNGHVH 398
> SPAC630.12
Length=422
Score = 33.5 bits (75), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 29/108 (26%), Positives = 45/108 (41%), Gaps = 12/108 (11%)
Query 77 SSKPAFIAILGDLVHAPVPALSQESLQDEVRAVRDQQAKDLKDVIDKPSKDVPVLVLPGN 136
S KP I+GDL+ ++E +D R + +V+D P + + PGN
Sbjct 90 SLKPDITFIMGDLMDTGREFATEEFKKDYFR---------MMNVLD-PKFTNKLEIYPGN 139
Query 137 HDV--GNAPTAASIADYEQLWGKDHFSFWFGGVKFVAANSSVLYDDTN 182
HD+ GN I +E L+G S G V + L ++ N
Sbjct 140 HDIGFGNHAIVKDIQRFESLFGPTSRSIDVGNHTLVIVDGIRLSNNVN 187
> At3g10150
Length=367
Score = 32.7 bits (73), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 31/107 (28%), Positives = 45/107 (42%), Gaps = 23/107 (21%)
Query 37 SSPFTFALIADPQFGLYRNNNSWEE--ERQQVAECIIAAASLSSK-PAFIAILGDLVHAP 93
SPF A+ AD FG ++W + Q V + +A L ++ P F+ LGD+V A
Sbjct 32 GSPFKIAIFADLHFG----EDTWTDWGPGQDVNSVNVMSAVLDAETPDFVVYLGDVVTAN 87
Query 94 VPALSQESL--QDEVRAVRDQQAKDLKDVIDKPSKDVPVLVLPGNHD 138
A+ SL + RD + +P L GNHD
Sbjct 88 NIAIQNASLFWDKAISPTRD--------------RGIPWATLFGNHD 120
> YDR182w
Length=491
Score = 32.0 bits (71), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 28/55 (50%), Gaps = 2/55 (3%)
Query 130 VLVLPGNHDVGNAPTA--ASIADYEQLWGKDHFSFWFGGVKFVAANSSVLYDDTN 182
V+ LPGNHD+G T +S+ + +G+ S G FV ++ L D TN
Sbjct 177 VMSLPGNHDIGFGDTVVESSLQRFSSYFGETSSSLDAGNHTFVLLDTISLSDKTN 231
> At3g52780
Length=427
Score = 32.0 bits (71), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 39/158 (24%), Positives = 58/158 (36%), Gaps = 37/158 (23%)
Query 129 PVLVLPGNHDVGNAPTAAS--IADYEQLW----------GKDHFSFWFGGVKFVAANSSV 176
P +V GNH++ P S Y + W ++SF GV + S
Sbjct 200 PWMVTHGNHELEKIPILHSNPFTAYNKRWRMPFEESGSSSNLYYSFNVYGVHIIMLGS-- 257
Query 177 LYDDTNAKELAEEQFEWIENELKDGMEKGAKQQFLLQHHQFYWDNWGEADDKTNTVEVRQ 236
Y D E EQ++W+EN LK K + H +Y N +K +VE+
Sbjct 258 -YTDF---EPGSEQYQWLENNLKKIDRKTTPWVVAVVHAPWYNSNEAHQGEK-ESVEM-- 310
Query 237 MRAHNPVSLFQLPKRWRERFQPLMKKYNVAASFHGHLH 274
+E + L+ K V F GH+H
Sbjct 311 ----------------KESMETLLYKARVDLVFAGHVH 332
> At2g30170
Length=283
Score = 30.4 bits (67), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 34/79 (43%), Gaps = 2/79 (2%)
Query 241 NPVSLFQLPKRWRERFQPLMKKYNVAASFHGHLHYDSLLQDGDTPQIVTSSCG--FATND 298
NP + L + QPL + +++ H H D + + G+ V+S G A D
Sbjct 23 NPSRVDFLCRCAPSEIQPLRPELSLSVGIHAIPHPDKVEKGGEDAFFVSSYRGGVMAVAD 82
Query 299 GRPGWRLVTVEESKFSHEF 317
G GW V+ S FS E
Sbjct 83 GVSGWAEQDVDPSLFSKEL 101
> 7298997
Length=418
Score = 30.4 bits (67), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 29/65 (44%), Gaps = 0/65 (0%)
Query 49 QFGLYRNNNSWEEERQQVAECIIAAASLSSKPAFIAILGDLVHAPVPALSQESLQDEVRA 108
Q G N +S+E E AE +I+ SLSS+ + ++ L H + R
Sbjct 169 QLGYMPNRDSFEREYDPTAEQLISNISLSSEDTEVDVMLKLAHVDIYTRRLRERARRKRM 228
Query 109 VRDQQ 113
VRD Q
Sbjct 229 VRDYQ 233
> 7292972
Length=649
Score = 29.6 bits (65), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 36/148 (24%), Positives = 53/148 (35%), Gaps = 9/148 (6%)
Query 178 YDDTNAKELAEEQFEWIENELKDGMEKGAKQQFLLQHHQFYWDNWGEADDKTNTVEVRQM 237
Y+DTNA A EQ W + + G F+ + GE DD VE
Sbjct 187 YEDTNAAPAAPEQPNWCLD-----YDDGNGDGFVQDNSSSTSQQAGEQDDDAEEVEGEDA 241
Query 238 RAHNPVSLFQLPKRWRERFQPLMKKYNVAASFHGHLHYDSLLQDGDTPQIVTSSCGFATN 297
S R + R + ++K A G + GD + V C + N
Sbjct 242 GGDCDASTSDGQPRKKRRREMIIKNATFMAGVPGVRQVSDQPRLGDLQRKVQDWCDWKKN 301
Query 298 DGRPGWRLVTVEE---SKFSHEFYPVKW 322
G PG + V+++ + S Y V W
Sbjct 302 -GFPGAQPVSMDRENIKRLSEIPYRVSW 328
Lambda K H
0.318 0.133 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7988630762
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40