bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2815_orf1
Length=185
Score E
Sequences producing significant alignments: (Bits) Value
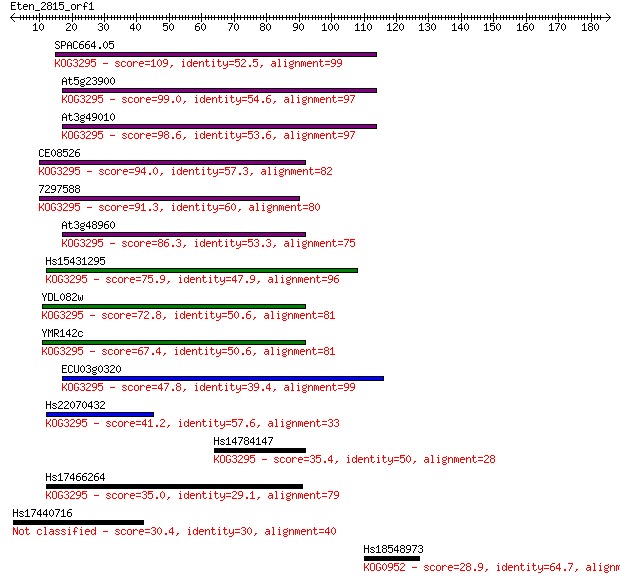
SPAC664.05 109 2e-24
At5g23900 99.0 4e-21
At3g49010 98.6 6e-21
CE08526 94.0 2e-19
7297588 91.3 1e-18
At3g48960 86.3 3e-17
Hs15431295 75.9 5e-14
YDL082w 72.8 4e-13
YMR142c 67.4 2e-11
ECU03g0320 47.8 1e-05
Hs22070432 41.2 0.001
Hs14784147 35.4 0.069
Hs17466264 35.0 0.085
Hs17440716 30.4 2.2
Hs18548973 28.9 5.3
> SPAC664.05
Length=208
Score = 109 bits (273), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 52/99 (52%), Positives = 65/99 (65%), Gaps = 10/99 (10%)
Query 15 AQLPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPLGLLRPAVRCPTQKYN 74
QLPN H K+WQR+V+TWF+QP RK RRR RQ KAAK+ RP+ +RPAV+ PT +YN
Sbjct 7 GQLPNAHFHKDWQRYVKTWFNQPGRKLRRRQARQTKAAKIAPRPVEAIRPAVKPPTIRYN 66
Query 75 IKLREGRGFTLEELKVSPDCCCCCCCCCCCTRPASLLGL 113
+K+R GRGFTLEELK R AS +G+
Sbjct 67 MKVRAGRGFTLEELK----------AAGVSRRVASTIGI 95
> At5g23900
Length=206
Score = 99.0 bits (245), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 53/102 (51%), Positives = 64/102 (62%), Gaps = 5/102 (4%)
Query 17 LPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPL-GLLRPAVRCPTQKYNI 75
+P+ H RK WQ +V+TWF+QPARK RRR+ RQKKA K+ RP G LRP V T KYN+
Sbjct 7 IPSSHFRKHWQNYVKTWFNQPARKTRRRVARQKKAVKIFPRPTSGPLRPVVHGQTLKYNM 66
Query 76 KLREGRGFTLEELKVS--PDCCCCCCCCCCCTRPA--SLLGL 113
K+R G+GFTLEELKV+ P R SL GL
Sbjct 67 KVRAGKGFTLEELKVAGIPKKLAPTIGISVDHRRKNRSLEGL 108
> At3g49010
Length=206
Score = 98.6 bits (244), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 52/102 (50%), Positives = 63/102 (61%), Gaps = 5/102 (4%)
Query 17 LPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPL-GLLRPAVRCPTQKYNI 75
+PN H +K WQ +V+TWF+QPARK RRR+ RQKKA K+ RP G LRP V T KYN+
Sbjct 7 IPNGHFKKHWQNYVKTWFNQPARKTRRRIARQKKAVKIFPRPTSGPLRPVVHGQTLKYNM 66
Query 76 KLREGRGFTLEELKVS--PDCCCCCCCCCCCTRPA--SLLGL 113
K+R G+GFTLEELK + P R SL GL
Sbjct 67 KVRTGKGFTLEELKAAGIPKKLAPTIGIAVDHRRKNRSLEGL 108
> CE08526
Length=207
Score = 94.0 bits (232), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 47/84 (55%), Positives = 58/84 (69%), Gaps = 2/84 (2%)
Query 10 MVSRSAQ-LPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPL-GLLRPAVR 67
M R Q L N H RK W + ++TWFDQPARK RRR RQ KA ++ RP+ GLLR VR
Sbjct 1 MAPRGNQMLGNAHFRKHWHKRIKTWFDQPARKLRRRQNRQAKAVEIAPRPVAGLLRSVVR 60
Query 68 CPTQKYNIKLREGRGFTLEELKVS 91
CP ++YN K R GRGF+L+ELK +
Sbjct 61 CPQKRYNTKTRLGRGFSLQELKAA 84
> 7297588
Length=218
Score = 91.3 bits (225), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 48/81 (59%), Positives = 55/81 (67%), Gaps = 1/81 (1%)
Query 10 MVSRSAQLPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPL-GLLRPAVRC 68
M + +PN H K WQR V+TWF+QPARK RR R KKA + RP G LRP VRC
Sbjct 1 MGKGNNMIPNQHYHKWWQRHVKTWFNQPARKVRRHANRVKKAKAVFPRPASGALRPVVRC 60
Query 69 PTQKYNIKLREGRGFTLEELK 89
PT +Y+ KLR GRGFTLEELK
Sbjct 61 PTIRYHTKLRAGRGFTLEELK 81
> At3g48960
Length=206
Score = 86.3 bits (212), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 40/76 (52%), Positives = 54/76 (71%), Gaps = 1/76 (1%)
Query 17 LPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPL-GLLRPAVRCPTQKYNI 75
+PN H +K+W+ +V+T F+QPA K RRR+ RQ KA K+ RP G +RP V T YN+
Sbjct 7 IPNGHFKKKWENYVKTSFNQPAMKTRRRIARQNKAVKIFPRPTAGPIRPVVHAQTLTYNM 66
Query 76 KLREGRGFTLEELKVS 91
K+R G+GFTLEELK +
Sbjct 67 KVRAGKGFTLEELKAA 82
> Hs15431295
Length=211
Score = 75.9 bits (185), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 46/97 (47%), Positives = 59/97 (60%), Gaps = 1/97 (1%)
Query 12 SRSAQLPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPL-GLLRPAVRCPT 70
SR+ + H K+WQR V TWF+QPARK RRR RQ KA ++ RP G +RP VRCPT
Sbjct 4 SRNGMVLKPHFHKDWQRRVATWFNQPARKIRRRKARQAKARRIAPRPASGPIRPIVRCPT 63
Query 71 QKYNIKLREGRGFTLEELKVSPDCCCCCCCCCCCTRP 107
+Y+ K+R GRGF+LEEL+V+ P
Sbjct 64 VRYHTKVRAGRGFSLEELRVAGIHKKVARTIGISVDP 100
> YDL082w
Length=199
Score = 72.8 bits (177), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 41/81 (50%), Positives = 52/81 (64%), Gaps = 0/81 (0%)
Query 11 VSRSAQLPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPLGLLRPAVRCPT 70
+S++ + H RK WQ V+ FDQ +K RR R +AAK+ RPL LLRP VR PT
Sbjct 3 ISKNLPILKNHFRKHWQERVKVHFDQAGKKVSRRNARATRAAKIAPRPLDLLRPVVRAPT 62
Query 71 QKYNIKLREGRGFTLEELKVS 91
KYN K+R GRGFTL E+K +
Sbjct 63 VKYNRKVRAGRGFTLAEVKAA 83
> YMR142c
Length=199
Score = 67.4 bits (163), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 41/81 (50%), Positives = 52/81 (64%), Gaps = 0/81 (0%)
Query 11 VSRSAQLPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPLGLLRPAVRCPT 70
+S++ + H RK WQ V+ FDQ +K RR R +AAK+ RPL LLRP VR PT
Sbjct 3 ISKNLPILKNHFRKHWQERVKVHFDQAGKKVSRRNARAARAAKIAPRPLDLLRPVVRAPT 62
Query 71 QKYNIKLREGRGFTLEELKVS 91
KYN K+R GRGFTL E+K +
Sbjct 63 VKYNRKVRAGRGFTLAEVKAA 83
> ECU03g0320
Length=163
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 39/100 (39%), Positives = 46/100 (46%), Gaps = 13/100 (13%)
Query 17 LPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQ-KKAAKLGVRPLGLLRPAVRCPTQKYNI 75
LPN H RK + +R D P K R +++ +KA L PL LRP VRCPT KYN
Sbjct 7 LPNNHFRKTSLK-IRIHHD-PETKARVMAEKKLRKAKALFPMPLKKLRPIVRCPTIKYNR 64
Query 76 KLREGRGFTLEELKVSPDCCCCCCCCCCCTRPASLLGLCC 115
R GRGFT E C R A LG+
Sbjct 65 NERLGRGFTAAE----------CEKAGLDYRHARRLGIAV 94
> Hs22070432
Length=119
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/33 (57%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 12 SRSAQLPNVHLRKEWQRFVRTWFDQPARKQRRR 44
SR+ + H K+WQR V TWF+QPARK RRR
Sbjct 4 SRNGMVLKPHFHKDWQRRVATWFNQPARKIRRR 36
> Hs14784147
Length=144
Score = 35.4 bits (80), Expect = 0.069, Method: Compositional matrix adjust.
Identities = 14/28 (50%), Positives = 21/28 (75%), Gaps = 0/28 (0%)
Query 64 PAVRCPTQKYNIKLREGRGFTLEELKVS 91
P VRCP +++ K + GRG +LEEL+V+
Sbjct 31 PIVRCPMLRHHYKAQAGRGLSLEELRVA 58
> Hs17466264
Length=161
Score = 35.0 bits (79), Expect = 0.085, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 33/80 (41%), Gaps = 24/80 (30%)
Query 12 SRSAQLPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPLGLLRPA-VRCPT 70
SR + H K WQ V TWF+ PA +RC
Sbjct 4 SRKGMIFKSHFNKHWQWCVATWFNHV-----------------------WTHPAHIRCCM 40
Query 71 QKYNIKLREGRGFTLEELKV 90
+Y+ K+ G+GF++EEL+V
Sbjct 41 VRYHTKVHAGQGFSMEELRV 60
> Hs17440716
Length=245
Score = 30.4 bits (67), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 12/40 (30%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 2 CCCFPQAKMVSRSAQLPNVHLRKEWQRFVRTWFDQPARKQ 41
C P+++ +S++A LPN+ WQ + D AR +
Sbjct 40 CEVKPESQHISKTAHLPNLSNEIRWQDSMEVGSDSEARSE 79
> Hs18548973
Length=731
Score = 28.9 bits (63), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 11/18 (61%), Positives = 12/18 (66%), Gaps = 1/18 (5%)
Query 110 LLGLCCRPWA-FPPEWPC 126
LLG+C R W FPP WP
Sbjct 13 LLGVCARGWGFFPPPWPA 30
Lambda K H
0.328 0.135 0.482
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2986559618
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40