bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2850_orf3
Length=225
Score E
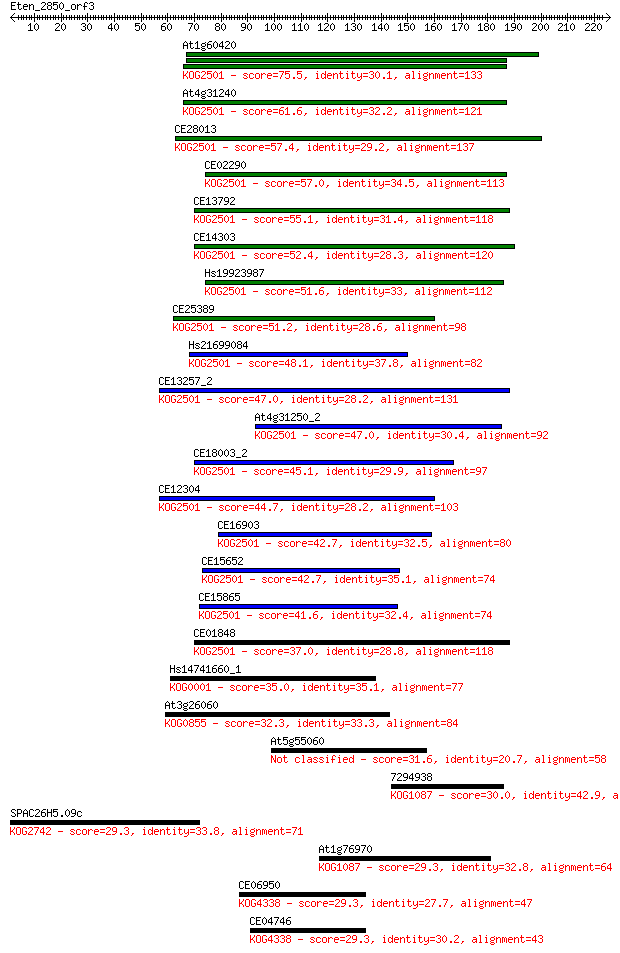
Sequences producing significant alignments: (Bits) Value
At1g60420 75.5 8e-14
At4g31240 61.6 1e-09
CE28013 57.4 2e-08
CE02290 57.0 3e-08
CE13792 55.1 1e-07
CE14303 52.4 8e-07
Hs19923987 51.6 1e-06
CE25389 51.2 2e-06
Hs21699084 48.1 1e-05
CE13257_2 47.0 3e-05
At4g31250_2 47.0 3e-05
CE18003_2 45.1 1e-04
CE12304 44.7 1e-04
CE16903 42.7 5e-04
CE15652 42.7 6e-04
CE15865 41.6 0.001
CE01848 37.0 0.029
Hs14741660_1 35.0 0.12
At3g26060 32.3 0.73
At5g55060 31.6 1.4
7294938 30.0 4.5
SPAC26H5.09c 29.3 5.9
At1g76970 29.3 6.8
CE06950 29.3 7.4
CE04746 29.3 7.5
> At1g60420
Length=578
Score = 75.5 bits (184), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 40/136 (29%), Positives = 72/136 (52%), Gaps = 20/136 (14%)
Query 67 NGNVVSQQHLIGKSVGLYFADGSSPKCSSFLPFLLQFYRTVNEGGSHQKIEVVFVSADKD 126
+G V L+GK++ +YF+ P C +F P L++ Y+ + E ++ E++F+S+D+D
Sbjct 352 DGAKVLVSDLVGKTILMYFSAHWCPPCRAFTPKLVEVYKQIKE--RNEAFELIFISSDRD 409
Query 127 ERAFQDHVKHMPWLVIDFNDPLRTILLRHFRVEKEASVPTQGQGPRAGVPSLVVVGSDG- 185
+ +F ++ MPWL + F DP + L + F+V G+P L +G G
Sbjct 410 QESFDEYYSQMPWLALPFGDPRKASLAKTFKV--------------GGIPMLAALGPTGQ 455
Query 186 ---RDAQFLQVSGGRD 198
++A+ L V+ G D
Sbjct 456 TVTKEARDLVVAHGAD 471
Score = 75.5 bits (184), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 40/120 (33%), Positives = 63/120 (52%), Gaps = 16/120 (13%)
Query 67 NGNVVSQQHLIGKSVGLYFADGSSPKCSSFLPFLLQFYRTVNEGGSHQKIEVVFVSADKD 126
+GN V L GK++GL F+ S KC+ P L++FY + E + + E+V +S + D
Sbjct 192 DGNKVPVSELEGKTIGLLFSVASYRKCTELTPKLVEFYTKLKE--NKEDFEIVLISLEDD 249
Query 127 ERAFQDHVKHMPWLVIDFNDPLRTILLRHFRVEKEASVPTQGQGPRAGVPSLVVVGSDGR 186
E +F K PWL + FND + L RHF + + +P+LV++G DG+
Sbjct 250 EESFNQDFKTKPWLALPFNDKSGSKLARHFML--------------STLPTLVILGPDGK 295
Score = 71.2 bits (173), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 43/122 (35%), Positives = 61/122 (50%), Gaps = 18/122 (14%)
Query 66 SNGNVVSQQHLIGKSVGLYFADGSSPKCSSFLPFLLQFYRTVNEGGSHQKIEVVFVSADK 125
++G V L+GK +GLYF+ C F P L++ Y NE S E+VFVS D+
Sbjct 31 NDGEQVKVDSLLGKKIGLYFSAAWCGPCQRFTPQLVEVY---NELSSKVGFEIVFVSGDE 87
Query 126 DERAFQDHVKHMPWLVIDFNDP-LRTILLRHFRVEKEASVPTQGQGPRAGVPSLVVVGSD 184
DE +F D+ + MPWL + F D R L F+V G+P+LV+V
Sbjct 88 DEESFGDYFRKMPWLAVPFTDSETRDRLDELFKVR--------------GIPNLVMVDDH 133
Query 185 GR 186
G+
Sbjct 134 GK 135
> At4g31240
Length=204
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 39/123 (31%), Positives = 59/123 (47%), Gaps = 19/123 (15%)
Query 66 SNGNVVSQQHLIGKSVGLYFADGSSPKCSSFLPFLLQFYRTVNEGGSHQK--IEVVFVSA 123
+N V L+GK++GLYF P SF L+ Y NE + K EV+ +S
Sbjct 3 TNVMQVLVSKLVGKTIGLYFGAHWCPPFRSFTSQLVDVY---NELATTDKGSFEVILIST 59
Query 124 DKDERAFQDHVKHMPWLVIDFNDPLRTILLRHFRVEKEASVPTQGQGPRAGVPSLVVVGS 183
D+D R F ++ +MPWL I + D R L R F V+ +P+LV++G
Sbjct 60 DRDSREFNINMTNMPWLAIPYEDRTRQDLCRIFNVKL--------------IPALVIIGP 105
Query 184 DGR 186
+ +
Sbjct 106 EEK 108
> CE28013
Length=179
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 40/139 (28%), Positives = 65/139 (46%), Gaps = 22/139 (15%)
Query 63 LKDSNGNVVSQQHLIGKSVGLYFADGSSPKCSSFLPFLLQFYRTVNEGGSHQKIEVVFVS 122
LK V +++ L GK V LYF+ G P C F P L++FY + + G + IEVVF S
Sbjct 39 LKQDLTEVPAEEALKGKVVALYFSAGWCPPCKQFTPKLVRFYHHLKKAG--KPIEVVFFS 96
Query 123 ADKDERAFQDHV--KHMPWLVIDFNDPLRTILLRHFRVEKEASVPTQGQGPRAGVPSLVV 180
D+ + +++ KH WL + + D + T F ++ +P L V
Sbjct 97 RDRSKADLEENFTEKHGDWLCVKYGDDILTRYQSKFEIK--------------TIPVLRV 142
Query 181 VGSDGRDAQFLQVSGGRDE 199
+ + G+ + V G+ E
Sbjct 143 INAAGK----MVVVDGKSE 157
> CE02290
Length=149
Score = 57.0 bits (136), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 39/115 (33%), Positives = 60/115 (52%), Gaps = 11/115 (9%)
Query 74 QHLIGKSVGLYFADGSSPKCSSFLPFLLQFYRTVNEGGSHQKIEVVFVSADKDERAFQDH 133
+HL GK VGLYF+ P C F P L +F+ + + H + EVVFVS D+++ +++
Sbjct 22 EHLKGKVVGLYFSASWCPPCRQFTPKLTRFFDEIRK--KHPEFEVVFVSRDREDGDLREY 79
Query 134 -VKHM-PWLVIDFNDPLRTILLRHFRVEKEASVPTQGQGPRAGVPSLVVVGSDGR 186
++HM W I F P LL + V+ ++P+ R P+ VV D R
Sbjct 80 FLEHMGAWTAIPFGTPRIQELLEQYEVK---TIPSM----RIVKPNGDVVVQDAR 127
> CE13792
Length=155
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 37/120 (30%), Positives = 56/120 (46%), Gaps = 21/120 (17%)
Query 70 VVSQQHLIGKSVGLYFADGSSPKCSSFLPFLLQFYRTVNEGGSHQKIEVVFVSADKDERA 129
V + + L GK VG YF+ P C F P L FY VNE + E+VFVS+D+ E
Sbjct 31 VDATEALAGKLVGFYFSAHWCPPCRGFTPILKDFYEEVNE-----EFEIVFVSSDRSESD 85
Query 130 FQDHVK--HMPWLVIDFNDPLRTILLRHFRVEKEASVPTQGQGPRAGVPSLVVVGSDGRD 187
+ ++K H W I + + L + V +G+P+L++V DG +
Sbjct 86 LKMYMKECHGDWYHIPHGNGAKQKLSTKYGV--------------SGIPALIIVKPDGTE 131
> CE14303
Length=122
Score = 52.4 bits (124), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 53/122 (43%), Gaps = 21/122 (17%)
Query 70 VVSQQHLIGKSVGLYFADGSSPKCSSFLPFLLQFYRTVNEGGSHQKIEVVFVSADKDERA 129
V + + L GK G YF+ P C F P L +FY V + E+VFVS+D E
Sbjct 7 VDATEALAGKIGGFYFSAHWCPPCCMFTPILKKFYEKV-----YDDFEIVFVSSDPSESG 61
Query 130 FQDHVK--HMPWLVIDFNDPLRTILLRHFRVEKEASVPTQGQGPRAGVPSLVVVGSDGRD 187
+ +++ H W I F + L + + G+P+LV+V DG +
Sbjct 62 LKKYMQECHGDWYYIPFGHEAKQKLCVKYEI--------------TGMPTLVIVKPDGTE 107
Query 188 AQ 189
+
Sbjct 108 VK 109
> Hs19923987
Length=212
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 37/117 (31%), Positives = 58/117 (49%), Gaps = 19/117 (16%)
Query 74 QHLIGKSVGLYFADGSSPKCSSFLPFLLQFY-RTVNEGGSHQ--KIEVVFVSADKDERAF 130
+ L + V L+F G+ P+C +F+P L F+ R +E + ++ +V+VS D E
Sbjct 28 RRLENRLVLLFFGAGACPQCQAFVPILKDFFVRLTDEFYVLRAAQLALVYVSQDSTEEQQ 87
Query 131 QDHVKHMP--WLVIDFNDPLRTILLRHFRVEKEASVPTQGQGPRAGVPSLVVVGSDG 185
+K MP WL + F D LR L R F VE+ +P++VV+ DG
Sbjct 88 DLFLKDMPKKWLFLPFEDDLRRDLGRQFSVER--------------LPAVVVLKPDG 130
> CE25389
Length=149
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/101 (27%), Positives = 54/101 (53%), Gaps = 5/101 (4%)
Query 62 SLKDSNGNVV-SQQHLIGKSVGLYFADGSSPKCSSFLPFLLQFYRTVNEGGSHQKIEVVF 120
+L++ G+ + +++HL GK +GLYF+ P C +F P L +F+ + + +H + E++F
Sbjct 9 TLENREGDELPAEEHLKGKIIGLYFSASWCPPCRAFTPKLKEFFEEIKK--THPEFEIIF 66
Query 121 VSADKDERAFQDHVKHM--PWLVIDFNDPLRTILLRHFRVE 159
VS D++ + K W I F L++ + V+
Sbjct 67 VSRDRNSSDLVTYFKEHQGEWTYIPFGSDKIMSLMQKYEVK 107
> Hs21699084
Length=135
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 38/85 (44%), Gaps = 3/85 (3%)
Query 68 GNVVSQQHLIGKSVGLYFADGSSPKCSSFLPFLLQFYRT-VNEGGSHQKIEVVFVSADKD 126
V ++ L K V LYFA F P L FY V E EVVFVSAD
Sbjct 16 ATVEAEAALQNKVVALYFAAARCAPSRDFTPLLCDFYTALVAEARRPAPFEVVFVSADGS 75
Query 127 ERAFQDHVK--HMPWLVIDFNDPLR 149
+ D ++ H WL + F+DP R
Sbjct 76 CQEMLDFMRELHGAWLALPFHDPYR 100
> CE13257_2
Length=745
Score = 47.0 bits (110), Expect = 3e-05, Method: Composition-based stats.
Identities = 37/133 (27%), Positives = 63/133 (47%), Gaps = 22/133 (16%)
Query 57 LFPEGSLKDSNGNVVSQQHLIGKSVGLYFADGSSPKCSSFLPFLLQFYRTVNEGGSHQKI 116
L P GS S++ L GK +GLY++ F P L QFY V+ +
Sbjct 614 LHPSGSY--------SERMLDGKVIGLYYSGYWCQPSRDFTPILAQFYSQVD-----KNF 660
Query 117 EVVFVSADKDERAFQDHVK--HMPWLVIDFNDPLRTILLRHFRVEKEASVPTQGQGPRAG 174
E++F+S+D+ E+ +++ H W + F+ P+ + L+ F + ++PT
Sbjct 661 EILFISSDRSEQEMNYYLQSSHGDWFHLPFDSPI-SKHLQQFNTKN--AIPTL----III 713
Query 175 VPSLVVVGSDGRD 187
P+ V+ DGRD
Sbjct 714 KPNGTVITVDGRD 726
> At4g31250_2
Length=281
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 28/92 (30%), Positives = 44/92 (47%), Gaps = 16/92 (17%)
Query 93 CSSFLPFLLQFYRTVNEGGSHQKIEVVFVSADKDERAFQDHVKHMPWLVIDFNDPLRTIL 152
C F P L++ Y + G +++E++FVS D D +F +H MPWL + FN L L
Sbjct 138 CKDFTPELIKLYENLQNRG--EELEIIFVSFDHDMTSFYEHFWCMPWLAVPFNLSLLNKL 195
Query 153 LRHFRVEKEASVPTQGQGPRAGVPSLVVVGSD 184
+ + + +PSLV + SD
Sbjct 196 RDKYGISR--------------IPSLVPLYSD 213
> CE18003_2
Length=144
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/99 (29%), Positives = 48/99 (48%), Gaps = 10/99 (10%)
Query 70 VVSQQHLIGKSVGLYFADGSSPKCSSFLPFLLQFYRTVNEGGSHQKIEVVFVSADKDERA 129
V + + L GK VG+YF+ C +F P L FY V + E+VF S+D+ E
Sbjct 19 VDANEALAGKIVGIYFSAHWCGPCRNFTPVLKDFYEEVQDD-----FEIVFASSDQSESD 73
Query 130 FQDHVK--HMPWLVIDFNDPLRTILLRHFRVEKEASVPT 166
+++++ H W I F + L + V +++PT
Sbjct 74 LKNYMEECHGNWYYIPFGNDAEEKLSTKYDV---STIPT 109
> CE12304
Length=174
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 50/106 (47%), Gaps = 5/106 (4%)
Query 57 LFPEGSLKDSNGNVV-SQQHLIGKSVGLYFADGSSPKCSSFLPFLLQFYRTVNEGGSHQK 115
F SL+ +G +V + +HL GK V LYF+ C F P + + Y+ + ++Q
Sbjct 5 FFSGTSLRLKDGTMVDAGEHLKGKIVVLYFSASWCGPCRQFTPIMKELYQQI--AATNQP 62
Query 116 IEVVFVSADKDERAFQDHVKH--MPWLVIDFNDPLRTILLRHFRVE 159
IEV+ +S D ++ + W V+ DP+ L + V+
Sbjct 63 IEVILLSRDYMRFQLDEYYESHGCSWGVVPLRDPIIEKCLEKYDVK 108
> CE16903
Length=151
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 39/81 (48%), Gaps = 3/81 (3%)
Query 79 KSVGLYFADGSSPKCSSFLPFLLQFYRTVNEGGSHQKIEVVFVSADKDERAFQDHVKH-M 137
K V LYF+ C F P L +FY + G ++IEVV VS D++ +++ H
Sbjct 28 KVVALYFSAMWCGSCRQFTPKLKRFYEALKAAG--KEIEVVLVSRDREAEDLLEYLGHGG 85
Query 138 PWLVIDFNDPLRTILLRHFRV 158
W+ I F D L+ + V
Sbjct 86 DWVAIPFGDERIQEYLKKYEV 106
> CE15652
Length=178
Score = 42.7 bits (99), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 42/77 (54%), Gaps = 4/77 (5%)
Query 73 QQHLIGKSVGLYFADGSSPKCSSFLPFLLQFYRTVNEGGSHQKIEVVFVSADKDERAFQD 132
+ + K+V +YF+ G C P + +FY V E + + +E+V+VS DK E A Q+
Sbjct 46 KDYFENKTVVVYFSAGWCGSCKFLTPKIKKFYNAVKESDAGKNLEIVWVSKDK-EAAHQE 104
Query 133 --HVKHMP-WLVIDFND 146
+ K++P W I F D
Sbjct 105 EYYEKNLPDWPYIPFGD 121
> CE15865
Length=777
Score = 41.6 bits (96), Expect = 0.001, Method: Composition-based stats.
Identities = 24/76 (31%), Positives = 38/76 (50%), Gaps = 7/76 (9%)
Query 72 SQQHLIGKSVGLYFADGSSPKCSSFLPFLLQFYRTVNEGGSHQKIEVVFVSADKDERAFQ 131
+++ L GK VGLYF+ P F P L QFY V + E++FVS+D + +
Sbjct 653 NERVLDGKVVGLYFSAHWCPPSRDFTPVLAQFYSQVED-----NFEILFVSSDNNTQEMN 707
Query 132 DHVK--HMPWLVIDFN 145
+++ H W + N
Sbjct 708 FYLQNFHGDWFHLPLN 723
> CE01848
Length=140
Score = 37.0 bits (84), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 34/120 (28%), Positives = 56/120 (46%), Gaps = 22/120 (18%)
Query 70 VVSQQHLIGKSVGLYFADGSSPKCSSFLPFLLQFYRTVNEGGSHQKIEVVFVSADKDERA 129
V + + L GK+VG YF+ P C F P L FY + EVVFVS D+ E
Sbjct 17 VDATEALAGKAVGFYFSAHWCPPCRGFTPILKDFYEE-----VEDEFEVVFVSFDRSESD 71
Query 130 FQDHV-KHMPWLVIDF-NDPLRTILLRHFRVEKEASVPTQGQGPRAGVPSLVVVGSDGRD 187
+ ++ +H W I + ND ++ + ++ +G+P+L++V DG +
Sbjct 72 LKMYMSEHGDWYHIPYGNDAIKELSTKY---------------GVSGIPALIIVKPDGTE 116
> Hs14741660_1
Length=629
Score = 35.0 bits (79), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 27/85 (31%), Positives = 44/85 (51%), Gaps = 19/85 (22%)
Query 61 GSLKDSNGNVV-------SQQHLIGKSVGL-YFADGSSPKCSSFLPFLLQFYRTVNEGGS 112
G+L SNGN+V ++Q L+ K L FA+G++ + SS L EG
Sbjct 372 GNLPSSNGNIVLPSEECVTEQSLLPKVGSLASFAEGNADEQSSGL-----------EGAC 420
Query 113 HQKIEVVFVSADKDERAFQDHVKHM 137
+E++ +ADK +A + H+KH+
Sbjct 421 KVNLELLLTNADKGLKAPEQHLKHV 445
> At3g26060
Length=216
Score = 32.3 bits (72), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 42/88 (47%), Gaps = 10/88 (11%)
Query 59 PEGSLKDSNGNVVSQQHLIGKSVGLYF--ADGSSPKCSSFLPFLLQFYRTVNEGGSHQKI 116
P+ +LKD NG VS + GK V LYF AD +P C+ Y + G+
Sbjct 76 PDFTLKDQNGKPVSLKKYKGKPVVLYFYPAD-ETPGCTKQACAFRDSYEKFKKAGA---- 130
Query 117 EVVFVSAD--KDERAFQDHVKHMPWLVI 142
EV+ +S D +AF K +P+ ++
Sbjct 131 EVIGISGDDSASHKAFASKYK-LPYTLL 157
> At5g55060
Length=645
Score = 31.6 bits (70), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 12/58 (20%), Positives = 31/58 (53%), Gaps = 0/58 (0%)
Query 99 FLLQFYRTVNEGGSHQKIEVVFVSADKDERAFQDHVKHMPWLVIDFNDPLRTILLRHF 156
F+++ + + +++ + + ++ R F + KH+PW+ +D N L++ LL +
Sbjct 212 FVVKLAEVIGSFTTPRRMALFWCRVVEELRRFWNEEKHIPWIPLDNNPDLKSCLLHQW 269
> 7294938
Length=543
Score = 30.0 bits (66), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 18/42 (42%), Positives = 21/42 (50%), Gaps = 3/42 (7%)
Query 144 FNDPLRTILLRHFRVEKEASVPTQGQGPRAGVPSLVVVGSDG 185
ND L + LRH R EK S QGQG PS V+ + G
Sbjct 291 INDELNNVFLRHQRYEKNRS---QGQGAGVTSPSAVLGAAMG 329
> SPAC26H5.09c
Length=369
Score = 29.3 bits (64), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 29/71 (40%), Gaps = 14/71 (19%)
Query 1 FPFHSHFLKMLPQYCSPYNAEKRRFNEEQILAAGGKEGAAAPAMPMPPAYRDMDLILFPE 60
F FH FLK LP + Y A +RR A E A A P Y +D +
Sbjct 16 FIFHYPFLKALPNHFEVYAAWERR--------ATSTESKARAAFPNVKVYTKLDEL---- 63
Query 61 GSLKDSNGNVV 71
L DSN +V
Sbjct 64 --LADSNIELV 72
> At1g76970
Length=387
Score = 29.3 bits (64), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 30/73 (41%), Gaps = 9/73 (12%)
Query 117 EVVFVSADKDERAFQDHVKHMPWLVID---------FNDPLRTILLRHFRVEKEASVPTQ 167
E V V + R +Q V + D ND L+ +L RH + SVP+
Sbjct 149 EEVIVDLVEQCRTYQRRVMTLVNTTTDEELLCQGLALNDNLQHVLQRHDDIANVGSVPSN 208
Query 168 GQGPRAGVPSLVV 180
G+ RA P +V
Sbjct 209 GRNTRAPPPVQIV 221
> CE06950
Length=1613
Score = 29.3 bits (64), Expect = 7.4, Method: Composition-based stats.
Identities = 13/47 (27%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 87 DGSSPKCSSFLPFLLQFYRTVNEGGSHQKIEVVFVSADKDERAFQDH 133
+ + P+ S FL L++ +RT + + E ++V ADK ++ +H
Sbjct 347 ENNMPETSHFLARLVRIFRTTSTSQLKEIHETLYVKADKKIQSLMEH 393
> CE04746
Length=1616
Score = 29.3 bits (64), Expect = 7.5, Method: Composition-based stats.
Identities = 13/43 (30%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 91 PKCSSFLPFLLQFYRTVNEGGSHQKIEVVFVSADKDERAFQDH 133
P+ S FL L++ +RT + + E ++V ADK ++ +H
Sbjct 351 PETSHFLARLVRIFRTTSTSQLKEIHETLYVKADKKIQSLMEH 393
Lambda K H
0.321 0.139 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4277266320
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40