bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2867_orf1
Length=202
Score E
Sequences producing significant alignments: (Bits) Value
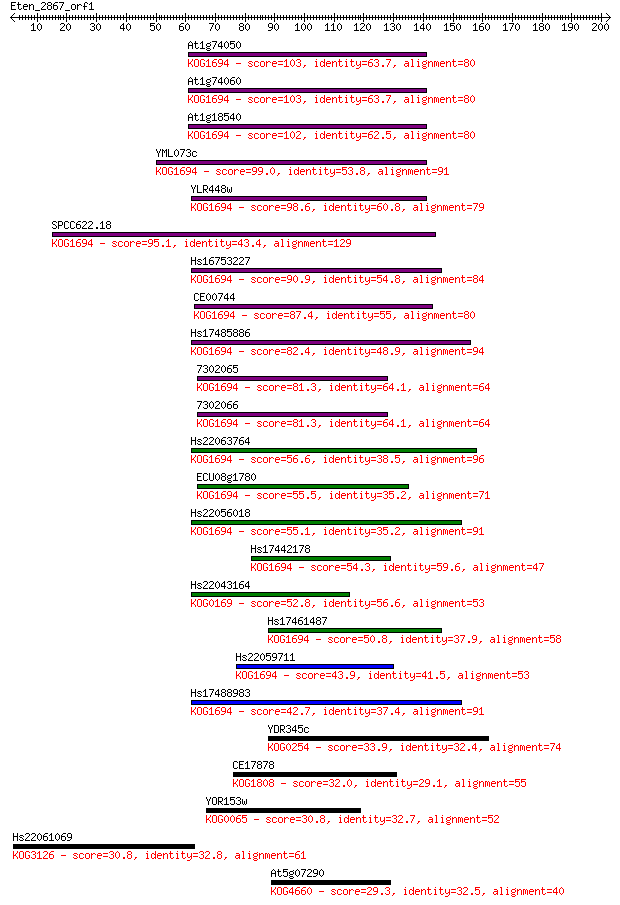
At1g74050 103 2e-22
At1g74060 103 2e-22
At1g18540 102 6e-22
YML073c 99.0 5e-21
YLR448w 98.6 7e-21
SPCC622.18 95.1 8e-20
Hs16753227 90.9 1e-18
CE00744 87.4 2e-17
Hs17485886 82.4 6e-16
7302065 81.3 1e-15
7302066 81.3 1e-15
Hs22063764 56.6 3e-08
ECU08g1780 55.5 7e-08
Hs22056018 55.1 9e-08
Hs17442178 54.3 1e-07
Hs22043164 52.8 5e-07
Hs17461487 50.8 2e-06
Hs22059711 43.9 2e-04
Hs17488983 42.7 5e-04
YDR345c 33.9 0.23
CE17878 32.0 0.93
YOR153w 30.8 1.7
Hs22061069 30.8 2.1
At5g07290 29.3 5.0
> At1g74050
Length=233
Score = 103 bits (258), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 51/80 (63%), Positives = 62/80 (77%), Gaps = 1/80 (1%)
Query 61 SKLRSSITPGTVLILLSSGFRGKRVVCLQQQQPSGLLLVTGPFAVNGVPLRRVNPRYVIA 120
+KLR+SITPGTVLI+L+ F+GKRVV L+Q SGLLLVTGPF +NGVPLRRVN YVI
Sbjct 83 TKLRASITPGTVLIILAGRFKGKRVVFLKQLA-SGLLLVTGPFKINGVPLRRVNQAYVIG 141
Query 121 TSSKINISEVDLSGVQQEMF 140
TS+K++IS V L + F
Sbjct 142 TSTKVDISGVTLDKFDDKYF 161
> At1g74060
Length=233
Score = 103 bits (258), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 51/80 (63%), Positives = 62/80 (77%), Gaps = 1/80 (1%)
Query 61 SKLRSSITPGTVLILLSSGFRGKRVVCLQQQQPSGLLLVTGPFAVNGVPLRRVNPRYVIA 120
+KLR+SITPGTVLI+L+ F+GKRVV L+Q SGLLLVTGPF +NGVPLRRVN YVI
Sbjct 83 AKLRASITPGTVLIILAGRFKGKRVVFLKQLA-SGLLLVTGPFKINGVPLRRVNQAYVIG 141
Query 121 TSSKINISEVDLSGVQQEMF 140
TS+K++IS V L + F
Sbjct 142 TSTKVDISGVTLDKFDDKYF 161
> At1g18540
Length=233
Score = 102 bits (253), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 50/80 (62%), Positives = 62/80 (77%), Gaps = 1/80 (1%)
Query 61 SKLRSSITPGTVLILLSSGFRGKRVVCLQQQQPSGLLLVTGPFAVNGVPLRRVNPRYVIA 120
+KL++SITPGTVLI+L+ F+GKRVV L+Q SGLLLVTGPF +NGVPLRRVN YVI
Sbjct 83 TKLKASITPGTVLIILAGRFKGKRVVFLKQLS-SGLLLVTGPFKINGVPLRRVNQAYVIG 141
Query 121 TSSKINISEVDLSGVQQEMF 140
TS+KI+IS V+ + F
Sbjct 142 TSTKIDISGVNTEKFDDKYF 161
> YML073c
Length=176
Score = 99.0 bits (245), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 49/91 (53%), Positives = 64/91 (70%), Gaps = 1/91 (1%)
Query 50 ALKRQAQGRSSSKLRSSITPGTVLILLSSGFRGKRVVCLQQQQPSGLLLVTGPFAVNGVP 109
ALK+ + KLR+S+ PGTVLILL+ FRGKRVV L+ + LL++GPF VNGVP
Sbjct 17 ALKKTRKAARPQKLRASLVPGTVLILLAGRFRGKRVVYLKHLE-DNTLLISGPFKVNGVP 75
Query 110 LRRVNPRYVIATSSKINISEVDLSGVQQEMF 140
LRRVN RYVIATS+K+++ V++ E F
Sbjct 76 LRRVNARYVIATSTKVSVEGVNVEKFNVEYF 106
> YLR448w
Length=176
Score = 98.6 bits (244), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 48/79 (60%), Positives = 59/79 (74%), Gaps = 1/79 (1%)
Query 62 KLRSSITPGTVLILLSSGFRGKRVVCLQQQQPSGLLLVTGPFAVNGVPLRRVNPRYVIAT 121
KLR+S+ PGTVLILL+ FRGKRVV L+ + LLVTGPF VNGVPLRRVN RYVIAT
Sbjct 29 KLRASLVPGTVLILLAGRFRGKRVVYLKHLE-DNTLLVTGPFKVNGVPLRRVNARYVIAT 87
Query 122 SSKINISEVDLSGVQQEMF 140
S+K+++ V++ E F
Sbjct 88 STKVSVEGVNVEKFNVEYF 106
> SPCC622.18
Length=195
Score = 95.1 bits (235), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 56/129 (43%), Positives = 79/129 (61%), Gaps = 10/129 (7%)
Query 15 VRSCGKMSAGKSAAAAAAAAGARRFRLRRKNKRAIALKRQAQGRSSSKLRSSITPGTVLI 74
V+ G + G+ A A A+ + R+N K+ + +KLR+S+ PGTV I
Sbjct 4 VKVNGAKNGGERMVLPAGEAAAKYYPAYREN----VPKKARKAVRPTKLRASLAPGTVCI 59
Query 75 LLSSGFRGKRVVCLQQQQPSGLLLVTGPFAVNGVPLRRVNPRYVIATSSKINISEVDLSG 134
LL+ FRGKRVV L Q + + L+VTGP+ VNGVP+RRVN RYVIATS+ ++D+SG
Sbjct 60 LLAGRFRGKRVVVLSQLEDT--LVVTGPYKVNGVPIRRVNHRYVIATSA----PKIDVSG 113
Query 135 VQQEMFVRS 143
V E F ++
Sbjct 114 VSVEKFTKA 122
> Hs16753227
Length=288
Score = 90.9 bits (224), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 46/84 (54%), Positives = 62/84 (73%), Gaps = 1/84 (1%)
Query 62 KLRSSITPGTVLILLSSGFRGKRVVCLQQQQPSGLLLVTGPFAVNGVPLRRVNPRYVIAT 121
KLR+SITPGT+LI+L+ RGKRVV L+Q SGLLLVTGP +N VPLRR + ++VIAT
Sbjct 139 KLRASITPGTILIILTGRHRGKRVVFLKQLA-SGLLLVTGPLVLNRVPLRRTHQKFVIAT 197
Query 122 SSKINISEVDLSGVQQEMFVRSKQ 145
S+KI+IS V + + + + K+
Sbjct 198 STKIDISNVKIPKHLTDAYFKKKK 221
> CE00744
Length=217
Score = 87.4 bits (215), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 44/81 (54%), Positives = 57/81 (70%), Gaps = 1/81 (1%)
Query 63 LRSSITPGTVLILLSSGFRGKRVVCLQQQQPSGLLLVTGPFAVNGVPLRRVNPRYVIATS 122
LR ++TPGTVLI+L+ +GKRVV L+Q SGLLLVTGP +NG PLRR+ +VIATS
Sbjct 67 LRKTLTPGTVLIVLAGRHKGKRVVFLKQLPQSGLLLVTGPHKINGFPLRRIGQAFVIATS 126
Query 123 SKINISEVDL-SGVQQEMFVR 142
K+N+S V + + E F R
Sbjct 127 LKVNVSGVKIPEHINDEYFKR 147
> Hs17485886
Length=293
Score = 82.4 bits (202), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 46/94 (48%), Positives = 64/94 (68%), Gaps = 1/94 (1%)
Query 62 KLRSSITPGTVLILLSSGFRGKRVVCLQQQQPSGLLLVTGPFAVNGVPLRRVNPRYVIAT 121
KLR+SITP T+LI+L+ RGKRVV L+Q SGLL VTGP N VPLRR + ++VIAT
Sbjct 144 KLRASITPWTILIILTGCHRGKRVVFLKQLG-SGLLPVTGPLVFNRVPLRRTHQKFVIAT 202
Query 122 SSKINISEVDLSGVQQEMFVRSKQQEARAAQAAQ 155
S+KI+IS V + ++ + +Q + QA++
Sbjct 203 STKIHISNVKIPKHLTGIYFKKQQLQKPRHQASE 236
> 7302065
Length=243
Score = 81.3 bits (199), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 41/64 (64%), Positives = 51/64 (79%), Gaps = 1/64 (1%)
Query 64 RSSITPGTVLILLSSGFRGKRVVCLQQQQPSGLLLVTGPFAVNGVPLRRVNPRYVIATSS 123
R ++TPGTVLILL+ +GKRVV L+ SGLLLVTGPFA+N PLRRV+ RYVI TSS
Sbjct 92 RRNLTPGTVLILLAGRHQGKRVVLLKVLA-SGLLLVTGPFALNSCPLRRVSQRYVIGTSS 150
Query 124 KINI 127
K+++
Sbjct 151 KVDL 154
> 7302066
Length=262
Score = 81.3 bits (199), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 41/64 (64%), Positives = 51/64 (79%), Gaps = 1/64 (1%)
Query 64 RSSITPGTVLILLSSGFRGKRVVCLQQQQPSGLLLVTGPFAVNGVPLRRVNPRYVIATSS 123
R ++TPGTVLILL+ +GKRVV L+ SGLLLVTGPFA+N PLRRV+ RYVI TSS
Sbjct 111 RRNLTPGTVLILLAGRHQGKRVVLLKVLA-SGLLLVTGPFALNSCPLRRVSQRYVIGTSS 169
Query 124 KINI 127
K+++
Sbjct 170 KVDL 173
> Hs22063764
Length=216
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 37/100 (37%), Positives = 52/100 (52%), Gaps = 10/100 (10%)
Query 62 KLRSSITPGTVLILLSSGFRGKRVVCLQQQQPSGLLLVTGPFAVNGVPLRRVNPRYVIAT 121
KLRSSIT T+LI+ + RG + P L TGP +N VPLRR + ++VIAT
Sbjct 123 KLRSSITSRTILIITTGRHRG------EGGFPEAAGLGTGPLVLNQVPLRRTHQKFVIAT 176
Query 122 SSKINISEVD----LSGVQQEMFVRSKQQEARAAQAAQQR 157
S+KI S V L+G + + K QE +++
Sbjct 177 STKIGSSNVKIAKRLTGAYFKKVWKPKHQEGETFDTEKEK 216
> ECU08g1780
Length=155
Score = 55.5 bits (132), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 48/72 (66%), Gaps = 2/72 (2%)
Query 64 RSSITPGTVLILLSSGFRGKRVVCLQQQQPSGLLLVTGPFAVNGVPLRRVNPRYVIATSS 123
R+ + G V+++L F +RVV L+ + + L GP ++NG+PL R++ RY++ATS+
Sbjct 24 RTDLVKGMVVVVLEGVFASRRVVYLKGLE-DNIALCAGPKSINGIPLFRIDERYLLATST 82
Query 124 KINIS-EVDLSG 134
++++ +VD+ G
Sbjct 83 VLDLNMDVDIDG 94
> Hs22056018
Length=201
Score = 55.1 bits (131), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 32/91 (35%), Positives = 47/91 (51%), Gaps = 23/91 (25%)
Query 62 KLRSSITPGTVLILLSSGFRGKRVVCLQQQQPSGLLLVTGPFAVNGVPLRRVNPRYVIAT 121
KLR+SITPGT+LI+L+ RGK V PLR+ + ++VIAT
Sbjct 75 KLRASITPGTILIILTGCHRGKEV-----------------------PLRKTHQKFVIAT 111
Query 122 SSKINISEVDLSGVQQEMFVRSKQQEARAAQ 152
S+KI+IS V + + + K Q+ R +
Sbjct 112 STKIDISNVKIPKHLTNAYFKKKLQKPRHQE 142
> Hs17442178
Length=221
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 28/47 (59%), Positives = 36/47 (76%), Gaps = 1/47 (2%)
Query 82 GKRVVCLQQQQPSGLLLVTGPFAVNGVPLRRVNPRYVIATSSKINIS 128
GKRV+ L+Q SG LLVTGP +N VPL+R + + VIATS+KI+IS
Sbjct 140 GKRVIFLKQLA-SGWLLVTGPLVLNQVPLQRTHQKSVIATSAKIDIS 185
> Hs22043164
Length=258
Score = 52.8 bits (125), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 30/53 (56%), Positives = 36/53 (67%), Gaps = 1/53 (1%)
Query 62 KLRSSITPGTVLILLSSGFRGKRVVCLQQQQPSGLLLVTGPFAVNGVPLRRVN 114
KLR +IT GTVLI+L GKRV L +Q SGLLLVTG +N VPL++ N
Sbjct 38 KLRGNITLGTVLIILPGHHTGKRVAFL-KQLGSGLLLVTGSLVLNQVPLQQKN 89
> Hs17461487
Length=221
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 37/58 (63%), Gaps = 0/58 (0%)
Query 88 LQQQQPSGLLLVTGPFAVNGVPLRRVNPRYVIATSSKINISEVDLSGVQQEMFVRSKQ 145
Q S LLL+TGP ++ VPL+R ++VIATS+KI++S V + +++ + K+
Sbjct 109 FSQHLASDLLLMTGPLVLDRVPLQRARWKFVIATSTKIDVSNVKIQKHLTDIYFKKKK 166
> Hs22059711
Length=138
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/53 (41%), Positives = 34/53 (64%), Gaps = 0/53 (0%)
Query 77 SSGFRGKRVVCLQQQQPSGLLLVTGPFAVNGVPLRRVNPRYVIATSSKINISE 129
SS K+++ +Q SGLLLVT P +N L + ++VIATS+KI+I++
Sbjct 21 SSSQHVKKILVALKQLSSGLLLVTEPIVLNQFLLHGTHQKFVIATSTKIDIND 73
> Hs17488983
Length=207
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 34/96 (35%), Positives = 50/96 (52%), Gaps = 9/96 (9%)
Query 62 KLRSSITPGTVLILLSSGFRGKRVVCLQQQQPSGLL----LVTGPFAVNGVPLRRVNPRY 117
KLR+SITP T+LI+L R KRVV L+Q SGL+ +GP + + + P
Sbjct 76 KLRASITPRTILIILIGHHRSKRVVFLKQLA-SGLVGTCDWTSGPQSSSST---KNTPEI 131
Query 118 VIATSSKINISEVDL-SGVQQEMFVRSKQQEARAAQ 152
TS++I+IS V + F + K Q+ R +
Sbjct 132 CHCTSTRIDISNVKTPKHLTDAYFTKKKLQKPRHQE 167
> YDR345c
Length=567
Score = 33.9 bits (76), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 39/84 (46%), Gaps = 10/84 (11%)
Query 88 LQQQQPSGLLLVTGPFAVNGVPLRRVNPRYVI---------ATSSKINISEVDLSGVQQE 138
+Q + P GL F + G+ +PRY++ A+ SK+N D +QQE
Sbjct 230 VQWRVPLGLCFAWALFMIGGMTFVPESPRYLVEAGQIDEARASLSKVNKVAPDHPFIQQE 289
Query 139 M-FVRSKQQEARAAQAAQQRRHFA 161
+ + + +EARAA +A F
Sbjct 290 LEVIEASVEEARAAGSASWGELFT 313
> CE17878
Length=2030
Score = 32.0 bits (71), Expect = 0.93, Method: Compositional matrix adjust.
Identities = 16/55 (29%), Positives = 27/55 (49%), Gaps = 0/55 (0%)
Query 76 LSSGFRGKRVVCLQQQQPSGLLLVTGPFAVNGVPLRRVNPRYVIATSSKINISEV 130
+ + FR K V + P + + G + G L +P+YVI + K N+SE+
Sbjct 683 IQAAFRFKVGVMPVPKHPENYVKIGGYYVEKGTQLTESDPKYVITKTVKGNLSEI 737
> YOR153w
Length=1511
Score = 30.8 bits (68), Expect = 1.7, Method: Composition-based stats.
Identities = 17/54 (31%), Positives = 29/54 (53%), Gaps = 4/54 (7%)
Query 67 ITPGTVLILLSSGFRGKRVV--CLQQQQPSGLLLVTGPFAVNGVPLRRVNPRYV 118
+ PGT+ L+ + GK + CL ++ G+ +TG VNG+P + PR +
Sbjct 895 VKPGTLTALMGASGAGKTTLLDCLAERVTMGV--ITGDILVNGIPRDKSFPRSI 946
> Hs22061069
Length=210
Score = 30.8 bits (68), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 33/62 (53%), Gaps = 1/62 (1%)
Query 2 VDKGSQLGLNRLEVRSCGKMSAGKSAAAAAAAAGARRFR-LRRKNKRAIALKRQAQGRSS 60
+KG LGL +L+VR+ + + G S + + A + F L K KR+I + + +SS
Sbjct 29 FNKGFGLGLVKLDVRTKSRSAVGFSTSGSFNADTGKAFEVLETKYKRSITGNKSGKIKSS 88
Query 61 SK 62
K
Sbjct 89 CK 90
> At5g07290
Length=907
Score = 29.3 bits (64), Expect = 5.0, Method: Composition-based stats.
Identities = 13/40 (32%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 89 QQQQPSGLLLVTGPFAVNGVPLRRVNPRYVIATSSKINIS 128
Q Q SG++ P VNGVP +R+ P + +S + ++
Sbjct 557 QHNQSSGMMWPNSPSRVNGVPSQRIPPVTAFSRASPLMVN 596
Lambda K H
0.318 0.127 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3551102874
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40