bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2921_orf2
Length=130
Score E
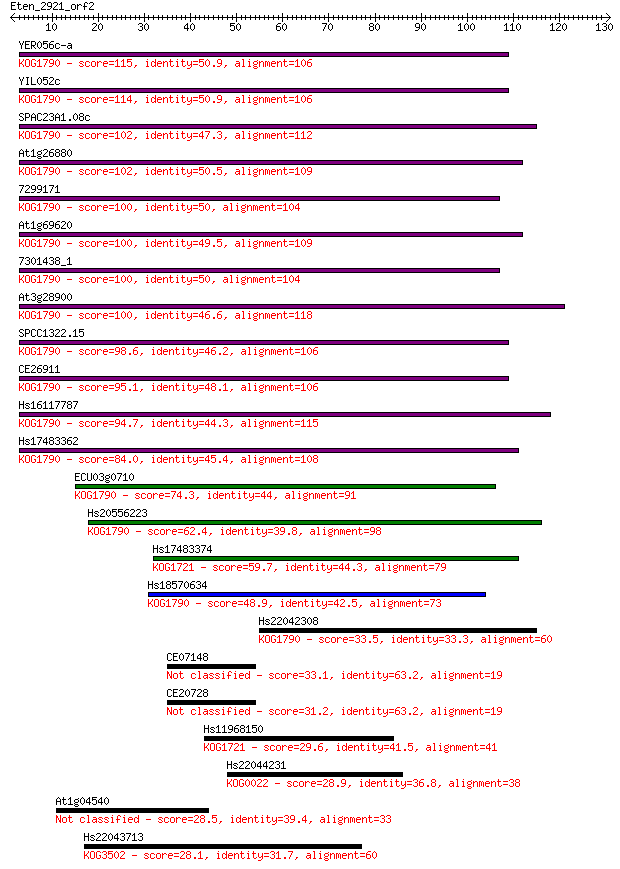
Sequences producing significant alignments: (Bits) Value
YER056c-a 115 3e-26
YIL052c 114 4e-26
SPAC23A1.08c 102 2e-22
At1g26880 102 2e-22
7299171 100 6e-22
At1g69620 100 7e-22
7301438_1 100 7e-22
At3g28900 100 9e-22
SPCC1322.15 98.6 3e-21
CE26911 95.1 3e-20
Hs16117787 94.7 4e-20
Hs17483362 84.0 6e-17
ECU03g0710 74.3 5e-14
Hs20556223 62.4 2e-10
Hs17483374 59.7 2e-09
Hs18570634 48.9 3e-06
Hs22042308 33.5 0.11
CE07148 33.1 0.14
CE20728 31.2 0.48
Hs11968150 29.6 1.6
Hs22044231 28.9 2.8
At1g04540 28.5 3.3
Hs22043713 28.1 4.2
> YER056c-a
Length=121
Score = 115 bits (287), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 54/106 (50%), Positives = 78/106 (73%), Gaps = 0/106 (0%)
Query 3 MSTRLVFRRHNHYNTASNRVRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPP 62
M+ R+ FRR N YNT SN+++ VKTPG + +V+K A+R +CG CG L GI RP
Sbjct 1 MAQRVTFRRRNPYNTRSNKIKVVKTPGGILRAQHVKKLATRPKCGDCGSALQGISTLRPR 60
Query 63 QFRLLKKRERTVNRAYGGTRCHSCVREKVLRAFLVEEQKCVKQVLQ 108
Q+ + K +TV+RAYGG+RC +CV+E+++RAFL+EEQK VK+V++
Sbjct 61 QYATVSKTHKTVSRAYGGSRCANCVKERIIRAFLIEEQKIVKKVVK 106
> YIL052c
Length=121
Score = 114 bits (286), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 54/106 (50%), Positives = 78/106 (73%), Gaps = 0/106 (0%)
Query 3 MSTRLVFRRHNHYNTASNRVRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPP 62
M+ R+ FRR N YNT SN+++ VKTPG + +V+K A+R +CG CG L GI RP
Sbjct 1 MAQRVTFRRRNPYNTRSNKIKVVKTPGGILRAQHVKKLATRPKCGDCGSALQGISTLRPR 60
Query 63 QFRLLKKRERTVNRAYGGTRCHSCVREKVLRAFLVEEQKCVKQVLQ 108
Q+ + K +TV+RAYGG+RC +CV+E+++RAFL+EEQK VK+V++
Sbjct 61 QYATVSKTHKTVSRAYGGSRCANCVKERIVRAFLIEEQKIVKKVVK 106
> SPAC23A1.08c
Length=112
Score = 102 bits (255), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 53/112 (47%), Positives = 73/112 (65%), Gaps = 0/112 (0%)
Query 3 MSTRLVFRRHNHYNTASNRVRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPP 62
M+ R+ +RR YNT SNR R +KTPG + YL+++K + RCG G L GIPA RP
Sbjct 1 MAQRVTYRRRLAYNTRSNRTRIIKTPGNNIRYLHIKKLGTIPRCGDTGVPLQGIPALRPR 60
Query 63 QFRLLKKRERTVNRAYGGTRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQK 114
+F L ++TV RAYGG + V+++++RAFL+EEQK VKQ L+ QK
Sbjct 61 EFARLSHNKKTVQRAYGGCLSANAVKDRIVRAFLIEEQKIVKQKLKQLSSQK 112
> At1g26880
Length=120
Score = 102 bits (254), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 55/111 (49%), Positives = 76/111 (68%), Gaps = 2/111 (1%)
Query 3 MSTRLVFRRHNHYNTASNRVRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPP 62
M RLV+R + Y T SN+ R VKTPG K+VY +K+AS +C G+ + GIP RP
Sbjct 1 MVQRLVYRSRHSYATKSNQHRIVKTPGGKLVYQTTKKRASGPKCPVTGKRIQGIPHLRPS 60
Query 63 QFRL--LKKRERTVNRAYGGTRCHSCVREKVLRAFLVEEQKCVKQVLQVKE 111
+++ L + RTVNRAYGG S VRE+++RAFLVEEQK VK+VL++++
Sbjct 61 EYKRSRLSRNRRTVNRAYGGVLSGSAVRERIIRAFLVEEQKIVKKVLKLQK 111
> 7299171
Length=168
Score = 100 bits (250), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 52/104 (50%), Positives = 68/104 (65%), Gaps = 0/104 (0%)
Query 3 MSTRLVFRRHNHYNTASNRVRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPP 62
M RL RR YNT SN+ R V+TPG ++VY V+K + RCG C L GI RP
Sbjct 1 MVQRLTLRRRLSYNTRSNKRRIVRTPGGRLVYQYVKKNPTVPRCGQCKEKLKGITPSRPS 60
Query 63 QFRLLKKRERTVNRAYGGTRCHSCVREKVLRAFLVEEQKCVKQV 106
+ + KR +TV+R YGG CHSC+RE+++RAFL+EEQK VK +
Sbjct 61 ERPRMSKRLKTVSRTYGGVLCHSCLRERIVRAFLIEEQKIVKAL 104
> At1g69620
Length=119
Score = 100 bits (249), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 54/111 (48%), Positives = 75/111 (67%), Gaps = 2/111 (1%)
Query 3 MSTRLVFRRHNHYNTASNRVRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPP 62
M RLV+R + Y T SN+ R VKTPG K+ Y +K+AS +C G+ + GIP RP
Sbjct 1 MVQRLVYRSRHSYATKSNQHRIVKTPGGKLTYQTTKKRASGPKCPVTGKRIQGIPHLRPT 60
Query 63 QFRL--LKKRERTVNRAYGGTRCHSCVREKVLRAFLVEEQKCVKQVLQVKE 111
+++ L + RTVNRAYGG S VRE+++RAFLVEEQK VK+VL++++
Sbjct 61 EYKRSRLSRNRRTVNRAYGGVLSGSAVRERIIRAFLVEEQKIVKKVLKLQK 111
> 7301438_1
Length=160
Score = 100 bits (249), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 52/104 (50%), Positives = 68/104 (65%), Gaps = 0/104 (0%)
Query 3 MSTRLVFRRHNHYNTASNRVRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPP 62
M RL RR YNT SN+ R V+TPG ++VY V+K + RCG C L GI RP
Sbjct 1 MVQRLTLRRRLSYNTRSNKRRIVRTPGGRLVYQYVKKNPTVPRCGQCKEKLHGITPSRPS 60
Query 63 QFRLLKKRERTVNRAYGGTRCHSCVREKVLRAFLVEEQKCVKQV 106
+ + KR +TV+R YGG CHSC+RE+++RAFL+EEQK VK +
Sbjct 61 ERPRMSKRLKTVSRTYGGVLCHSCLRERIVRAFLIEEQKIVKAL 104
> At3g28900
Length=120
Score = 100 bits (248), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 55/120 (45%), Positives = 79/120 (65%), Gaps = 2/120 (1%)
Query 3 MSTRLVFRRHNHYNTASNRVRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPP 62
M RLV+R + Y T SN+ R VKTPG K+ Y K+AS +C G+ + GIP RP
Sbjct 1 MVQRLVYRSRHSYATKSNQHRIVKTPGGKLTYQTTNKRASGPKCPVTGKRIQGIPHLRPA 60
Query 63 QFRL--LKKRERTVNRAYGGTRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQKRDEVKT 120
+++ L + ERTVNRAYGG VRE+++RAFLVEEQK VK+VL++++ +++ K+
Sbjct 61 EYKRSRLARNERTVNRAYGGVLSGVAVRERIVRAFLVEEQKIVKKVLKLQKAKEKTAPKS 120
> SPCC1322.15
Length=111
Score = 98.6 bits (244), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 49/106 (46%), Positives = 70/106 (66%), Gaps = 0/106 (0%)
Query 3 MSTRLVFRRHNHYNTASNRVRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPP 62
M+ R+ +RR YNT SN+ R +KTPG + YL+++K + RCG G L GIPA RP
Sbjct 1 MAQRVTYRRRLAYNTRSNKTRIIKTPGNNIRYLHIKKLGTIPRCGDTGVPLQGIPALRPR 60
Query 63 QFRLLKKRERTVNRAYGGTRCHSCVREKVLRAFLVEEQKCVKQVLQ 108
+F L ++ V RAYGG + V+++++RAFL+EEQK VKQ L+
Sbjct 61 EFARLSHNQKKVQRAYGGCLSANAVKDRIVRAFLIEEQKIVKQKLK 106
> CE26911
Length=110
Score = 95.1 bits (235), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 51/106 (48%), Positives = 68/106 (64%), Gaps = 0/106 (0%)
Query 3 MSTRLVFRRHNHYNTASNRVRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPP 62
MS R+ +RR YNT SN+ R VKTPG ++V ++K+ +C G L GI RP
Sbjct 1 MSLRVTYRRRLSYNTTSNKKRLVKTPGGRLVVQYIKKRGQIPKCRDTGVKLHGITPARPI 60
Query 63 QFRLLKKRERTVNRAYGGTRCHSCVREKVLRAFLVEEQKCVKQVLQ 108
RLLK+ ERTV RAYGG + V+E++ RAFLVEEQK V +V++
Sbjct 61 ALRLLKRNERTVTRAYGGCLSPNAVKERITRAFLVEEQKIVNKVIK 106
> Hs16117787
Length=117
Score = 94.7 bits (234), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 51/117 (43%), Positives = 75/117 (64%), Gaps = 2/117 (1%)
Query 3 MSTRLVFRRHNHYNTASNRVRRVKTPGAKVVYLNVRK--QASRQRCGGCGRLLPGIPARR 60
M RL +RR YNTASN+ R +TPG ++VYL +K +A + CG C L G+ A R
Sbjct 1 MVQRLTYRRRLSYNTASNKTRLSRTPGNRIVYLYTKKVGKAPKSACGVCPGRLRGVRAVR 60
Query 61 PPQFRLLKKRERTVNRAYGGTRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQKRDE 117
P L K ++ V+RAYGG+ C CVR+++ RAFL+EEQK V +VL+ + + ++ +
Sbjct 61 PKVLMRLSKTKKHVSRAYGGSMCAKCVRDRIKRAFLIEEQKIVVKVLKAQAQSQKAK 117
> Hs17483362
Length=117
Score = 84.0 bits (206), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 49/110 (44%), Positives = 68/110 (61%), Gaps = 2/110 (1%)
Query 3 MSTRLVFRRHNHYNTASNRVRRVKTPGAKVVYLNVRK--QASRQRCGGCGRLLPGIPARR 60
M RL ++ YNTASN+ R +TPG ++VYL +K +A R G C L G+ A R
Sbjct 1 MVQRLTYQCRLSYNTASNKTRLPRTPGNRIVYLYTKKVGKAPRSASGMCPGRLRGVRAVR 60
Query 61 PPQFRLLKKRERTVNRAYGGTRCHSCVREKVLRAFLVEEQKCVKQVLQVK 110
P L KR++ V+RAYGG+ C CVR ++ RAFL+ EQK V +VL+ +
Sbjct 61 PKVLMKLSKRKKHVSRAYGGSMCAKCVRGRIKRAFLIGEQKIVVKVLKAQ 110
> ECU03g0710
Length=110
Score = 74.3 bits (181), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 40/91 (43%), Positives = 53/91 (58%), Gaps = 0/91 (0%)
Query 15 YNTASNRVRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTV 74
Y T SNR R+V+TP K+V V+K + + RC C +L I RP +F K R V
Sbjct 12 YKTRSNRRRKVRTPSGKLVNRRVKKHSKKHRCHECNAILGSIARMRPAEFSRQKVSARRV 71
Query 75 NRAYGGTRCHSCVREKVLRAFLVEEQKCVKQ 105
NR YG T C CVREK++ AFL E++ V +
Sbjct 72 NRPYGATTCGRCVREKIISAFLGNEERIVME 102
> Hs20556223
Length=131
Score = 62.4 bits (150), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 39/100 (39%), Positives = 55/100 (55%), Gaps = 2/100 (2%)
Query 18 ASNRVRRVKTPGAKVVYLNVRK--QASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVN 75
A N+ R +TPG +VYL +K +A + CG C L G+ A RP L + V
Sbjct 30 AFNKTRLPRTPGNGIVYLYTKKVGKAPKSACGVCPGRLRGVLAVRPKVLMRLSNTRKHVR 89
Query 76 RAYGGTRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQKR 115
RA GG C CV +++ AFL+EEQK V +VL+ R ++
Sbjct 90 RACGGLMCAKCVCDRIKGAFLIEEQKIVVKVLKAHARGQK 129
> Hs17483374
Length=580
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 35/81 (43%), Positives = 49/81 (60%), Gaps = 2/81 (2%)
Query 32 VVYLNVRK--QASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVRE 89
+ YL +K +AS+ CG C L G+ A R L K + V+RAYGG+ C CVR
Sbjct 484 IEYLYTKKVGKASKSACGMCPGRLRGVRAVRLKVLMKLSKTNKHVSRAYGGSMCAKCVRG 543
Query 90 KVLRAFLVEEQKCVKQVLQVK 110
++ RAFL+EEQK V +VL+ +
Sbjct 544 RIKRAFLIEEQKIVVKVLKAQ 564
> Hs18570634
Length=277
Score = 48.9 bits (115), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 31/75 (41%), Positives = 46/75 (61%), Gaps = 2/75 (2%)
Query 31 KVVYLNVRK--QASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVR 88
++V+L +K +A + CG L G+ A RP L K ++ V+RAYGG+ C VR
Sbjct 29 RIVHLYTKKVGKAPKSACGVYPGRLRGVRAVRPKVLMRLSKTKKHVSRAYGGSMCAKRVR 88
Query 89 EKVLRAFLVEEQKCV 103
++V RAFL++EQK V
Sbjct 89 DRVKRAFLIKEQKIV 103
> Hs22042308
Length=102
Score = 33.5 bits (75), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 35/60 (58%), Gaps = 1/60 (1%)
Query 55 GIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQK 114
GI A RP + ++ + E+T + C C +K+ AFL++++K V +VL+ +ER K
Sbjct 4 GISAVRP-KVLMIGQYEKTCQQGLRCPMCAKCACDKLKGAFLIKKEKIVSKVLKAQERIK 62
> CE07148
Length=890
Score = 33.1 bits (74), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 12/19 (63%), Positives = 14/19 (73%), Gaps = 0/19 (0%)
Query 35 LNVRKQASRQRCGGCGRLL 53
+ VR QASR CGGCGR+
Sbjct 708 MEVRDQASRAECGGCGRVF 726
> CE20728
Length=805
Score = 31.2 bits (69), Expect = 0.48, Method: Composition-based stats.
Identities = 12/19 (63%), Positives = 14/19 (73%), Gaps = 0/19 (0%)
Query 35 LNVRKQASRQRCGGCGRLL 53
+ VR QASR CGGCGR+
Sbjct 623 MEVRDQASRAECGGCGRVF 641
> Hs11968150
Length=711
Score = 29.6 bits (65), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 20/41 (48%), Gaps = 6/41 (14%)
Query 43 RQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRC 83
R RCG CG+ L A R L +R T +R YG T C
Sbjct 406 RHRCGQCGKGLSSKTALR------LHERTHTGDRPYGCTEC 440
> Hs22044231
Length=312
Score = 28.9 bits (63), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 19/40 (47%), Gaps = 2/40 (5%)
Query 48 GCGRLLPGIPAR--RPPQFRLLKKRERTVNRAYGGTRCHS 85
GCG+ PG P LL+ E+TV R + CH+
Sbjct 212 GCGKYAPGSPLTCCWAQALVLLRGSEKTVTRTFQLVTCHT 251
> At1g04540
Length=601
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 18/33 (54%), Gaps = 0/33 (0%)
Query 11 RHNHYNTASNRVRRVKTPGAKVVYLNVRKQASR 43
R Y T++N RR PG + V L VR+ + R
Sbjct 106 RRPGYRTSNNEYRRTPPPGMRFVALQVRRTSGR 138
> Hs22043713
Length=106
Score = 28.1 bits (61), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 30/64 (46%), Gaps = 7/64 (10%)
Query 17 TASNRVRRVKTPGAKVVYLNVRKQA----SRQRCGGCGRLLPGIPARRPPQFRLLKKRER 72
T+ + +R VK P V + NV+ + R C +LPG+P L++K E
Sbjct 38 TSRSIIRNVKGP---VGFFNVKGRWVRPHGRWECLNASLILPGVPKCESLSIDLIQKEEI 94
Query 73 TVNR 76
T N+
Sbjct 95 TFNQ 98
Lambda K H
0.322 0.134 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1246445644
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40