bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2929_orf1
Length=135
Score E
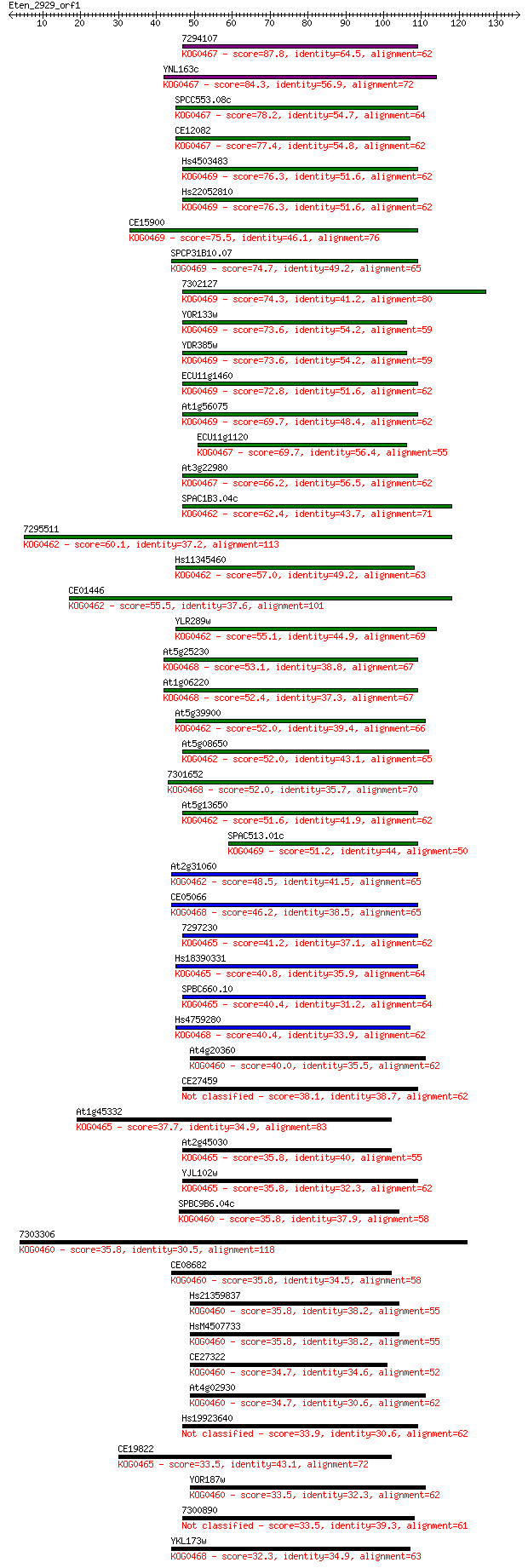
Sequences producing significant alignments: (Bits) Value
7294107 87.8 5e-18
YNL163c 84.3 5e-17
SPCC553.08c 78.2 5e-15
CE12082 77.4 7e-15
Hs4503483 76.3 2e-14
Hs22052810 76.3 2e-14
CE15900 75.5 2e-14
SPCP31B10.07 74.7 5e-14
7302127 74.3 6e-14
YOR133w 73.6 1e-13
YDR385w 73.6 1e-13
ECU11g1460 72.8 2e-13
At1g56075 69.7 1e-12
ECU11g1120 69.7 2e-12
At3g22980 66.2 1e-11
SPAC1B3.04c 62.4 2e-10
7295511 60.1 1e-09
Hs11345460 57.0 1e-08
CE01446 55.5 3e-08
YLR289w 55.1 4e-08
At5g25230 53.1 1e-07
At1g06220 52.4 2e-07
At5g39900 52.0 3e-07
At5g08650 52.0 3e-07
7301652 52.0 3e-07
At5g13650 51.6 4e-07
SPAC513.01c 51.2 5e-07
At2g31060 48.5 3e-06
CE05066 46.2 2e-05
7297230 41.2 5e-04
Hs18390331 40.8 7e-04
SPBC660.10 40.4 9e-04
Hs4759280 40.4 0.001
At4g20360 40.0 0.001
CE27459 38.1 0.005
At1g45332 37.7 0.006
At2g45030 35.8 0.021
YJL102w 35.8 0.023
SPBC9B6.04c 35.8 0.025
7303306 35.8 0.025
CE08682 35.8 0.026
Hs21359837 35.8 0.026
HsM4507733 35.8 0.026
CE27322 34.7 0.048
At4g02930 34.7 0.051
Hs19923640 33.9 0.099
CE19822 33.5 0.10
YOR187w 33.5 0.13
7300890 33.5 0.13
YKL173w 32.3 0.27
> 7294107
Length=860
Score = 87.8 bits (216), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 40/62 (64%), Positives = 51/62 (82%), Gaps = 0/62 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
VRNI ILAHVDHGKT+L+D L+A+NG I++ A KLR+LD+R DEQ R IT+K+S +SL
Sbjct 19 VRNICILAHVDHGKTTLADSLVASNGIISQRMAGKLRYLDNRSDEQERGITMKSSSISLY 78
Query 107 YQ 108
YQ
Sbjct 79 YQ 80
> YNL163c
Length=1110
Score = 84.3 bits (207), Expect = 5e-17, Method: Composition-based stats.
Identities = 41/74 (55%), Positives = 57/74 (77%), Gaps = 2/74 (2%)
Query 42 SSPEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKAS 101
+ P +RNI I+AHVDHGKTSLSD LLA+NG I++ A K+RFLD+R DEQ R IT+++S
Sbjct 14 NDPSCIRNICIVAHVDHGKTSLSDSLLASNGIISQRLAGKIRFLDARPDEQLRGITMESS 73
Query 102 VVSLLYQ--RRPQG 113
+SL ++ R+ +G
Sbjct 74 AISLYFRVLRKQEG 87
> SPCC553.08c
Length=1000
Score = 78.2 bits (191), Expect = 5e-15, Method: Composition-based stats.
Identities = 35/64 (54%), Positives = 49/64 (76%), Gaps = 0/64 (0%)
Query 45 EFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVS 104
E +RN ++LAHVDHGKT+L+D LLA+NG I+ A +RFLD R+DE R IT+K+S +S
Sbjct 17 ENIRNFTLLAHVDHGKTTLADSLLASNGIISSKLAGTVRFLDFREDEITRGITMKSSAIS 76
Query 105 LLYQ 108
L ++
Sbjct 77 LFFK 80
> CE12082
Length=906
Score = 77.4 bits (189), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 34/62 (54%), Positives = 48/62 (77%), Gaps = 0/62 (0%)
Query 45 EFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVS 104
E +RN+ ++AHVDHGKTS +D L++ N I+ A KLR++DSR+DEQ R IT+K+S +S
Sbjct 18 EHIRNVCLVAHVDHGKTSFADSLVSANAVISSRMAGKLRYMDSREDEQTRGITMKSSGIS 77
Query 105 LL 106
LL
Sbjct 78 LL 79
> Hs4503483
Length=858
Score = 76.3 bits (186), Expect = 2e-14, Method: Composition-based stats.
Identities = 32/62 (51%), Positives = 47/62 (75%), Gaps = 0/62 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RN+S++AHVDHGK++L+D L+ G I A A + RF D+R+DEQ R ITIK++ +SL
Sbjct 19 IRNMSVIAHVDHGKSTLTDSLVCKAGIIASARAGETRFTDTRKDEQERCITIKSTAISLF 78
Query 107 YQ 108
Y+
Sbjct 79 YE 80
> Hs22052810
Length=846
Score = 76.3 bits (186), Expect = 2e-14, Method: Composition-based stats.
Identities = 32/62 (51%), Positives = 47/62 (75%), Gaps = 0/62 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RN+S++AHVDHGK++L+D L+ G I A A + RF D+R+DEQ R ITIK++ +SL
Sbjct 7 IRNMSVIAHVDHGKSTLTDSLVCKAGIIASARAGETRFTDTRKDEQERCITIKSTAISLF 66
Query 107 YQ 108
Y+
Sbjct 67 YE 68
> CE15900
Length=852
Score = 75.5 bits (184), Expect = 2e-14, Method: Composition-based stats.
Identities = 35/76 (46%), Positives = 53/76 (69%), Gaps = 6/76 (7%)
Query 33 AQMARKLNFSSPEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQ 92
A M RK N +RN+S++AHVDHGK++L+D L++ G I + A + RF D+R+DEQ
Sbjct 11 ALMDRKRN------IRNMSVIAHVDHGKSTLTDSLVSKAGIIAGSKAGETRFTDTRKDEQ 64
Query 93 NRQITIKASVVSLLYQ 108
R ITIK++ +SL ++
Sbjct 65 ERCITIKSTAISLFFE 80
> SPCP31B10.07
Length=842
Score = 74.7 bits (182), Expect = 5e-14, Method: Composition-based stats.
Identities = 32/65 (49%), Positives = 47/65 (72%), Gaps = 0/65 (0%)
Query 44 PEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVV 103
P VRN+S++AHVDHGK++L+D L+ G I+ A A RF+D+R DEQ R +TIK++ +
Sbjct 16 PSNVRNMSVIAHVDHGKSTLTDSLVQKAGIISAAKAGDARFMDTRADEQERGVTIKSTAI 75
Query 104 SLLYQ 108
SL +
Sbjct 76 SLFAE 80
> 7302127
Length=832
Score = 74.3 bits (181), Expect = 6e-14, Method: Composition-based stats.
Identities = 33/82 (40%), Positives = 57/82 (69%), Gaps = 2/82 (2%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RN+S++AHVDHGK++L+D L++ G I A A + RF D+R+DEQ R ITIK++ +S+
Sbjct 7 IRNMSVIAHVDHGKSTLTDSLVSKAGIIAGAKAGETRFTDTRKDEQERCITIKSTAISMY 66
Query 107 YQRRPQGP--LANPRKAQQETK 126
++ + + +P + ++E K
Sbjct 67 FEVEEKDLVFITHPDQREKECK 88
> YOR133w
Length=842
Score = 73.6 bits (179), Expect = 1e-13, Method: Composition-based stats.
Identities = 32/59 (54%), Positives = 46/59 (77%), Gaps = 0/59 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSL 105
VRN+S++AHVDHGK++L+D L+ G I+ A A + RF D+R+DEQ R ITIK++ +SL
Sbjct 19 VRNMSVIAHVDHGKSTLTDSLVQRAGIISAAKAGEARFTDTRKDEQERGITIKSTAISL 77
> YDR385w
Length=842
Score = 73.6 bits (179), Expect = 1e-13, Method: Composition-based stats.
Identities = 32/59 (54%), Positives = 46/59 (77%), Gaps = 0/59 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSL 105
VRN+S++AHVDHGK++L+D L+ G I+ A A + RF D+R+DEQ R ITIK++ +SL
Sbjct 19 VRNMSVIAHVDHGKSTLTDSLVQRAGIISAAKAGEARFTDTRKDEQERGITIKSTAISL 77
> ECU11g1460
Length=850
Score = 72.8 bits (177), Expect = 2e-13, Method: Composition-based stats.
Identities = 32/62 (51%), Positives = 48/62 (77%), Gaps = 1/62 (1%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RNIS++AHVDHGK++L+DCL+ +++ S R++DSR+DEQ R ITIK+S +SL
Sbjct 19 IRNISVIAHVDHGKSTLTDCLVIKAKIVSKDSGGG-RYMDSREDEQQRGITIKSSAISLH 77
Query 107 YQ 108
+Q
Sbjct 78 FQ 79
> At1g56075
Length=855
Score = 69.7 bits (169), Expect = 1e-12, Method: Composition-based stats.
Identities = 30/62 (48%), Positives = 45/62 (72%), Gaps = 0/62 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RN+S++AHVDHGK++L+D L+A G I + A +R D+R DE R ITIK++ +SL
Sbjct 31 IRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRADEAERGITIKSTGISLY 90
Query 107 YQ 108
Y+
Sbjct 91 YE 92
> ECU11g1120
Length=678
Score = 69.7 bits (169), Expect = 2e-12, Method: Composition-based stats.
Identities = 31/55 (56%), Positives = 42/55 (76%), Gaps = 0/55 (0%)
Query 51 SILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSL 105
S++AH+DHGKTSL D L+A+ G I+ A +RFLD+R+DEQ R IT+K V+SL
Sbjct 10 SVVAHIDHGKTSLIDSLVASQGRISRTLAGSIRFLDTREDEQARGITLKLGVISL 64
> At3g22980
Length=1015
Score = 66.2 bits (160), Expect = 1e-11, Method: Composition-based stats.
Identities = 35/64 (54%), Positives = 47/64 (73%), Gaps = 2/64 (3%)
Query 47 VRNISILAHVDHGKTSLSDCLLATN--GFITEASAAKLRFLDSRQDEQNRQITIKASVVS 104
VRNI ILAHVDHGKT+L+D L+A++ G + A KLRF+D +EQ R IT+K+S +S
Sbjct 9 VRNICILAHVDHGKTTLADHLIASSGGGVLHPRLAGKLRFMDYLDEEQRRAITMKSSSIS 68
Query 105 LLYQ 108
L Y+
Sbjct 69 LKYK 72
> SPAC1B3.04c
Length=646
Score = 62.4 bits (150), Expect = 2e-10, Method: Composition-based stats.
Identities = 31/71 (43%), Positives = 46/71 (64%), Gaps = 1/71 (1%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
VRN +++AH+DHGK++LSDC+L G I E + + +FLD + E+ R IT+KA S++
Sbjct 58 VRNWAVIAHIDHGKSTLSDCILKLTGVINEHN-FRNQFLDKLEVERRRGITVKAQTCSMI 116
Query 107 YQRRPQGPLAN 117
Y Q L N
Sbjct 117 YYYHGQSYLLN 127
> 7295511
Length=679
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 42/117 (35%), Positives = 60/117 (51%), Gaps = 5/117 (4%)
Query 5 QSARLGARGGSGFPGELKLTGCVAAS----AAAQMARKLNFSSPEFVRNISILAHVDHGK 60
SA L AR S L T V + A + R+ E +RN SI+AHVDHGK
Sbjct 53 HSAWLVARSKSLLVRNLSTTNQVKGETEEPSQADLLREFAHMPVERIRNFSIIAHVDHGK 112
Query 61 TSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLLYQRRPQGPLAN 117
++L+D LL G I + + + LD+ Q E+ R IT+KA S+ ++ + Q L N
Sbjct 113 STLADRLLELTGAIAR-NGGQHQVLDNLQVERERGITVKAQTASIFHRHKGQLYLLN 168
> Hs11345460
Length=669
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 31/63 (49%), Positives = 40/63 (63%), Gaps = 1/63 (1%)
Query 45 EFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVS 104
E +RN SI+AHVDHGK++L+D LL G I + K + LD Q E+ R IT+KA S
Sbjct 66 ENIRNFSIVAHVDHGKSTLTDRLLELTGTIDKTKNNK-QVLDKLQVERERGITVKAQTAS 124
Query 105 LLY 107
L Y
Sbjct 125 LFY 127
> CE01446
Length=645
Score = 55.5 bits (132), Expect = 3e-08, Method: Composition-based stats.
Identities = 38/103 (36%), Positives = 58/103 (56%), Gaps = 10/103 (9%)
Query 17 FPGELKLTGCVAASAAAQMARKLNFS--SPEFVRNISILAHVDHGKTSLSDCLLATNGFI 74
F G+L+++ C S A + +N S +P+ +RN I+AHVDHGK++L+D LL G +
Sbjct 11 FIGKLRVS-CKHYSTAGDPTKLVNLSEFTPDKIRNFGIVAHVDHGKSTLADRLLEMCGAV 69
Query 75 TEASAAKLRFLDSRQDEQNRQITIKASVVSLLYQRRPQGPLAN 117
+ + LD Q E+ R IT+KA +L R +G L N
Sbjct 70 ---PPGQKQMLDKLQVERERGITVKAQTAAL----RHRGYLLN 105
> YLR289w
Length=645
Score = 55.1 bits (131), Expect = 4e-08, Method: Composition-based stats.
Identities = 31/69 (44%), Positives = 42/69 (60%), Gaps = 1/69 (1%)
Query 45 EFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVS 104
E RN SI+AHVDHGK++LSD LL I + +A + LD + E+ R ITIKA S
Sbjct 44 ENYRNFSIVAHVDHGKSTLSDRLLEITHVI-DPNARNKQVLDKLEVERERGITIKAQTCS 102
Query 105 LLYQRRPQG 113
+ Y+ + G
Sbjct 103 MFYKDKRTG 111
> At5g25230
Length=973
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 26/70 (37%), Positives = 43/70 (61%), Gaps = 3/70 (4%)
Query 42 SSPEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAK---LRFLDSRQDEQNRQITI 98
S+P VRN++++ H+ HGKT D L+ ++ +A +R+ D+R DEQ R I+I
Sbjct 119 SNPALVRNVALVGHLQHGKTVFMDMLVEQTHRMSTFNAENDKHMRYTDTRVDEQERNISI 178
Query 99 KASVVSLLYQ 108
KA +SL+ +
Sbjct 179 KAVPMSLVLE 188
> At1g06220
Length=987
Score = 52.4 bits (124), Expect = 2e-07, Method: Composition-based stats.
Identities = 25/70 (35%), Positives = 43/70 (61%), Gaps = 3/70 (4%)
Query 42 SSPEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAK---LRFLDSRQDEQNRQITI 98
S+P VRN++++ H+ HGKT D L+ ++ +A +++ D+R DEQ R I+I
Sbjct 133 SNPALVRNVALVGHLQHGKTVFMDMLVEQTHHMSTFNAKNEKHMKYTDTRVDEQERNISI 192
Query 99 KASVVSLLYQ 108
KA +SL+ +
Sbjct 193 KAVPMSLVLE 202
> At5g39900
Length=661
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 42/66 (63%), Gaps = 3/66 (4%)
Query 45 EFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVS 104
E +RN SI+AH+DHGK++L+D L+ G I + + ++LD Q E R IT+KA +
Sbjct 64 EKIRNFSIIAHIDHGKSTLADRLMELTGTIKKGH-GQPQYLDKLQRE--RGITVKAQTAT 120
Query 105 LLYQRR 110
+ Y+ +
Sbjct 121 MFYENK 126
> At5g08650
Length=675
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 28/67 (41%), Positives = 43/67 (64%), Gaps = 3/67 (4%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIK--ASVVS 104
+RN SI+AH+DHGK++L+D LL G + + K +FLD+ E+ R ITIK A+ +
Sbjct 80 IRNFSIIAHIDHGKSTLADKLLQVTGTV-QNRDMKEQFLDNMDLERERGITIKLQAARMR 138
Query 105 LLYQRRP 111
+Y+ P
Sbjct 139 YVYEDTP 145
> 7301652
Length=975
Score = 52.0 bits (123), Expect = 3e-07, Method: Composition-based stats.
Identities = 25/71 (35%), Positives = 42/71 (59%), Gaps = 1/71 (1%)
Query 43 SPEFVRNISILAHVDHGKTSLSDCLL-ATNGFITEASAAKLRFLDSRQDEQNRQITIKAS 101
+P +RN++++ H+ HGKT+ DCL+ T+ +LR+ D+ EQ R +IKA+
Sbjct 127 TPPLIRNVALVGHLHHGKTTFVDCLIRQTHPQFETMEERQLRYTDTLFTEQERGCSIKAT 186
Query 102 VVSLLYQRRPQ 112
V+L+ Q Q
Sbjct 187 PVTLVLQDVKQ 197
> At5g13650
Length=609
Score = 51.6 bits (122), Expect = 4e-07, Method: Composition-based stats.
Identities = 26/62 (41%), Positives = 37/62 (59%), Gaps = 0/62 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
VRNI+I+AHVDHGKT+L D +L + + R +DS E+ R ITI + S+
Sbjct 16 VRNIAIVAHVDHGKTTLVDSMLRQAKVFRDNQVMQERIMDSNDLERERGITILSKNTSIT 75
Query 107 YQ 108
Y+
Sbjct 76 YK 77
> SPAC513.01c
Length=812
Score = 51.2 bits (121), Expect = 5e-07, Method: Composition-based stats.
Identities = 22/50 (44%), Positives = 34/50 (68%), Gaps = 0/50 (0%)
Query 59 GKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLLYQ 108
GK++L+D L+ G I+ A A RF+D+R DEQ R +TIK++ +SL +
Sbjct 1 GKSTLTDSLVQKAGIISAAKAGDARFMDTRADEQERGVTIKSTAISLFAE 50
> At2g31060
Length=664
Score = 48.5 bits (114), Expect = 3e-06, Method: Composition-based stats.
Identities = 27/65 (41%), Positives = 39/65 (60%), Gaps = 4/65 (6%)
Query 44 PEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVV 103
P +RN++++AHVDHGKT+L D LL G A R +DS E+ R ITI + V
Sbjct 56 PNRLRNVAVIAHVDHGKTTLMDRLLRQCG----ADIPHERAMDSINLERERGITISSKVT 111
Query 104 SLLYQ 108
S+ ++
Sbjct 112 SIFWK 116
> CE05066
Length=974
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 25/66 (37%), Positives = 37/66 (56%), Gaps = 1/66 (1%)
Query 44 PEFVRNISILAHVDHGKTSLSDCLL-ATNGFITEASAAKLRFLDSRQDEQNRQITIKASV 102
P +RN++I H+ HGKT+ DCL+ T+ A A RF D E+ R +IK+
Sbjct 129 PHIMRNVAIAGHLHHGKTTFLDCLMEQTHPEFYRAEDADARFTDILFIEKQRGCSIKSQP 188
Query 103 VSLLYQ 108
VS++ Q
Sbjct 189 VSIVAQ 194
> 7297230
Length=729
Score = 41.2 bits (95), Expect = 5e-04, Method: Composition-based stats.
Identities = 23/67 (34%), Positives = 39/67 (58%), Gaps = 5/67 (7%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLR-----FLDSRQDEQNRQITIKAS 101
+RNI I AH+D GKT+L++ +L G I E + + +DS + E+ R ITI+++
Sbjct 26 IRNIGISAHIDSGKTTLTERILFYTGRIAEMHEVRGKDNVGATMDSMELERQRGITIQSA 85
Query 102 VVSLLYQ 108
L++
Sbjct 86 ATYTLWK 92
> Hs18390331
Length=751
Score = 40.8 bits (94), Expect = 7e-04, Method: Composition-based stats.
Identities = 23/69 (33%), Positives = 40/69 (57%), Gaps = 5/69 (7%)
Query 45 EFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLR-----FLDSRQDEQNRQITIK 99
E +RNI I AH+D GKT+L++ +L G I + K + +DS + E+ R ITI+
Sbjct 44 EKIRNIGISAHIDSGKTTLTERVLYYTGRIAKMHEVKGKDGVGAVMDSMELERQRGITIQ 103
Query 100 ASVVSLLYQ 108
++ +++
Sbjct 104 SAATYTMWK 112
> SPBC660.10
Length=813
Score = 40.4 bits (93), Expect = 9e-04, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 37/66 (56%), Gaps = 2/66 (3%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKL--RFLDSRQDEQNRQITIKASVVS 104
+RN+ I+AH+D GKT+L++ +L GF + +D E+ R ITI ++ +S
Sbjct 28 IRNVGIIAHIDAGKTTLTEKMLYYGGFTSHFGNVDTGDTVMDYLPAERQRGITINSAAIS 87
Query 105 LLYQRR 110
++ +
Sbjct 88 FTWRNQ 93
> Hs4759280
Length=972
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 36/63 (57%), Gaps = 1/63 (1%)
Query 45 EFVRNISILAHVDHGKTSLSDCLL-ATNGFITEASAAKLRFLDSRQDEQNRQITIKASVV 103
E +RN+++ H+ HGKT DCL+ T+ I + L + D EQ R + IK++ V
Sbjct 127 ELIRNVTLCGHLHHGKTCFVDCLIEQTHPEIRKRYDQDLCYTDILFTEQERGVGIKSTPV 186
Query 104 SLL 106
+++
Sbjct 187 TVV 189
> At4g20360
Length=476
Score = 40.0 bits (92), Expect = 0.001, Method: Composition-based stats.
Identities = 22/62 (35%), Positives = 32/62 (51%), Gaps = 0/62 (0%)
Query 49 NISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLLYQ 108
NI + HVDHGKT+L+ L I + A K +D+ +E+ R ITI + V +
Sbjct 81 NIGTIGHVDHGKTTLTAALTMALASIGSSVAKKYDEIDAAPEERARGITINTATVEYETE 140
Query 109 RR 110
R
Sbjct 141 NR 142
> CE27459
Length=689
Score = 38.1 bits (87), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 39/67 (58%), Gaps = 8/67 (11%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAK-----LRFLDSRQDEQNRQITIKAS 101
+RNI ++AHVD GKT++++ LL G I A FLD E+ R IT++++
Sbjct 25 LRNIGVIAHVDAGKTTVTERLLYLAGAIHVAGHVDKGNTVTDFLDI---ERERGITVQSA 81
Query 102 VVSLLYQ 108
V+L ++
Sbjct 82 AVNLDWK 88
> At1g45332
Length=754
Score = 37.7 bits (86), Expect = 0.006, Method: Composition-based stats.
Identities = 29/92 (31%), Positives = 49/92 (53%), Gaps = 9/92 (9%)
Query 19 GELKLTGCVAASAAAQMARKLN----FSSPEFVRNISILAHVDHGKTSLSDCLLATNGFI 74
G+ +L +A AA++A+ S + +RNI I AH+D GKT+L++ +L G I
Sbjct 33 GDFQLIRHFSAGTAARVAKDEKEPWWKESMDKLRNIGISAHIDSGKTTLTERVLFYTGRI 92
Query 75 TEASAAKLR-----FLDSRQDEQNRQITIKAS 101
E + R +DS E+ + ITI+++
Sbjct 93 HEIHEVRGRDGVGAKMDSMDLEREKGITIQSA 124
> At2g45030
Length=754
Score = 35.8 bits (81), Expect = 0.021, Method: Composition-based stats.
Identities = 22/60 (36%), Positives = 35/60 (58%), Gaps = 5/60 (8%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLR-----FLDSRQDEQNRQITIKAS 101
+RNI I AH+D GKT+L++ +L G I E + R +DS E+ + ITI+++
Sbjct 65 LRNIGISAHIDSGKTTLTERVLFYTGRIHEIHEVRGRDGVGAKMDSMDLEREKGITIQSA 124
> YJL102w
Length=819
Score = 35.8 bits (81), Expect = 0.023, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 35/64 (54%), Gaps = 2/64 (3%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKL--RFLDSRQDEQNRQITIKASVVS 104
VRNI I+AH+D GKT+ ++ +L G D + E++R ITI+++ +S
Sbjct 41 VRNIGIIAHIDAGKTTTTERMLYYAGISKHIGDVDTGDTITDFLEQERSRGITIQSAAIS 100
Query 105 LLYQ 108
++
Sbjct 101 FPWR 104
> SPBC9B6.04c
Length=439
Score = 35.8 bits (81), Expect = 0.025, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 33/63 (52%), Gaps = 5/63 (7%)
Query 46 FVR-----NISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKA 100
FVR NI + HVDHGKT+L+ + + +AS +D +E+ R ITI +
Sbjct 47 FVRKKPHVNIGTIGHVDHGKTTLTAAITKCLSDLGQASFMDYSQIDKAPEEKARGITISS 106
Query 101 SVV 103
+ V
Sbjct 107 AHV 109
> 7303306
Length=489
Score = 35.8 bits (81), Expect = 0.025, Method: Composition-based stats.
Identities = 36/130 (27%), Positives = 55/130 (42%), Gaps = 12/130 (9%)
Query 4 LQSARL----GARGGSGFPGELKLTGCVAASAAAQMARKLNFSSPE---FVR-----NIS 51
+ SAR+ GA S + + G +AA M + L + E F R N+
Sbjct 27 MSSARMLRMAGATATSTACKDWQRNGLLAAPRTGNMQQYLREYANEKKVFERTKPHCNVG 86
Query 52 ILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLLYQRRP 111
+ HVDHGKT+L+ + A + K +D+ +E+ R ITI + V + R
Sbjct 87 TIGHVDHGKTTLTAAITKVLADKQLAESKKYNEIDNAPEEKARGITINVAHVEYQTETRH 146
Query 112 QGPLANPRKA 121
G P A
Sbjct 147 YGHTDCPGHA 156
> CE08682
Length=453
Score = 35.8 bits (81), Expect = 0.026, Method: Composition-based stats.
Identities = 20/61 (32%), Positives = 32/61 (52%), Gaps = 6/61 (9%)
Query 44 PEFVRNISILAHVDHGKTSLSDCLL---ATNGFITEASAAKLRFLDSRQDEQNRQITIKA 100
P+ N+ + H+DHGKT+L+ + A GF A K +D ++E+ R ITI
Sbjct 42 PKINVNVGTIGHIDHGKTTLTSAITRVQAKKGF---AKHIKFDEIDKGKEEKKRGITINV 98
Query 101 S 101
+
Sbjct 99 A 99
> Hs21359837
Length=452
Score = 35.8 bits (81), Expect = 0.026, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 32/58 (55%), Gaps = 6/58 (10%)
Query 49 NISILAHVDHGKTSLSDCLLATNGFITEASAAKLRF---LDSRQDEQNRQITIKASVV 103
N+ + HVDHGKT+L+ A + E AK + +D+ +E+ R ITI A+ V
Sbjct 59 NVGTIGHVDHGKTTLT---AAITKILAEGGGAKFKKYEEIDNAPEERARGITINAAHV 113
> HsM4507733
Length=452
Score = 35.8 bits (81), Expect = 0.026, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 32/58 (55%), Gaps = 6/58 (10%)
Query 49 NISILAHVDHGKTSLSDCLLATNGFITEASAAKLRF---LDSRQDEQNRQITIKASVV 103
N+ + HVDHGKT+L+ A + E AK + +D+ +E+ R ITI A+ V
Sbjct 59 NVGTIGHVDHGKTTLT---AAITKILAEGGGAKFKKYEEIDNAPEERARGITINAAHV 113
> CE27322
Length=496
Score = 34.7 bits (78), Expect = 0.048, Method: Composition-based stats.
Identities = 18/52 (34%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 49 NISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKA 100
N+ + HVDHGKT+L+ + A K +D+ +E+ R ITI A
Sbjct 52 NVGTIGHVDHGKTTLTSAITKILATSKGAKYRKYEDIDNAPEEKARGITINA 103
> At4g02930
Length=454
Score = 34.7 bits (78), Expect = 0.051, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 30/62 (48%), Gaps = 0/62 (0%)
Query 49 NISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLLYQ 108
N+ + HVDHGKT+L+ + +A A +D +E+ R ITI + V
Sbjct 69 NVGTIGHVDHGKTTLTAAITKVLAEEGKAKAIAFDEIDKAPEEKKRGITIATAHVEYETA 128
Query 109 RR 110
+R
Sbjct 129 KR 130
> Hs19923640
Length=779
Score = 33.9 bits (76), Expect = 0.099, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 37/67 (55%), Gaps = 8/67 (11%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFI-----TEASAAKLRFLDSRQDEQNRQITIKAS 101
+RNI I+AH+D GKT+ ++ +L +G+ + F+ E+ R ITI+++
Sbjct 70 IRNIGIMAHIDAGKTTTTERILYYSGYTRSLGDVDDGDTVTDFM---AQERERGITIQSA 126
Query 102 VVSLLYQ 108
V+ ++
Sbjct 127 AVTFDWK 133
> CE19822
Length=750
Score = 33.5 bits (75), Expect = 0.10, Method: Composition-based stats.
Identities = 31/99 (31%), Positives = 47/99 (47%), Gaps = 30/99 (30%)
Query 30 SAAAQMARKLN-----FSSPEF--------------VRNISILAHVDHGKTSLSDCLLAT 70
SA AQ R+LN F+S E +RNI I AH+D GKT++++ +L
Sbjct 8 SAVAQSRRQLNNVLRRFASNEAPSSVVVPGVRPIERIRNIGISAHIDSGKTTVTERILYY 67
Query 71 NGFI--------TEASAAKLRFLDSRQDEQNRQITIKAS 101
G I + A + F+D E+ R ITI+++
Sbjct 68 AGRIDSMHEVRGKDDVGATMDFMDL---ERQRGITIQSA 103
> YOR187w
Length=437
Score = 33.5 bits (75), Expect = 0.13, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 30/62 (48%), Gaps = 0/62 (0%)
Query 49 NISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLLYQ 108
NI + HVDHGKT+L+ + T A+ +D +E+ R ITI + V
Sbjct 50 NIGTIGHVDHGKTTLTAAITKTLAAKGGANFLDYAAIDKAPEERARGITISTAHVEYETA 109
Query 109 RR 110
+R
Sbjct 110 KR 111
> 7300890
Length=1093
Score = 33.5 bits (75), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 34/64 (53%), Gaps = 4/64 (6%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLR---FLDSRQDEQNRQITIKASVV 103
+RNI ILAH+D GKT+ ++ +L G T A R D E+ R ITI +S V
Sbjct 33 IRNIGILAHIDAGKTTTTERMLFYAGK-TRALGEVHRGNTVTDYLTQERERGITICSSAV 91
Query 104 SLLY 107
+ +
Sbjct 92 TFSW 95
> YKL173w
Length=1008
Score = 32.3 bits (72), Expect = 0.27, Method: Composition-based stats.
Identities = 22/70 (31%), Positives = 39/70 (55%), Gaps = 7/70 (10%)
Query 44 PEFVRNISILAHVDHGKTSLSDCL-LATNGFITEASA------AKLRFLDSRQDEQNRQI 96
PE + N+ ++ + GKTSL D L + ++ I + S LR+LD+ + E +R +
Sbjct 130 PERIINVGVIGPLHSGKTSLMDLLVIDSHKRIPDMSKNVELGWKPLRYLDNLKQEIDRGL 189
Query 97 TIKASVVSLL 106
+IK + +LL
Sbjct 190 SIKLNGSTLL 199
Lambda K H
0.316 0.129 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1388795348
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40