bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2961_orf3
Length=192
Score E
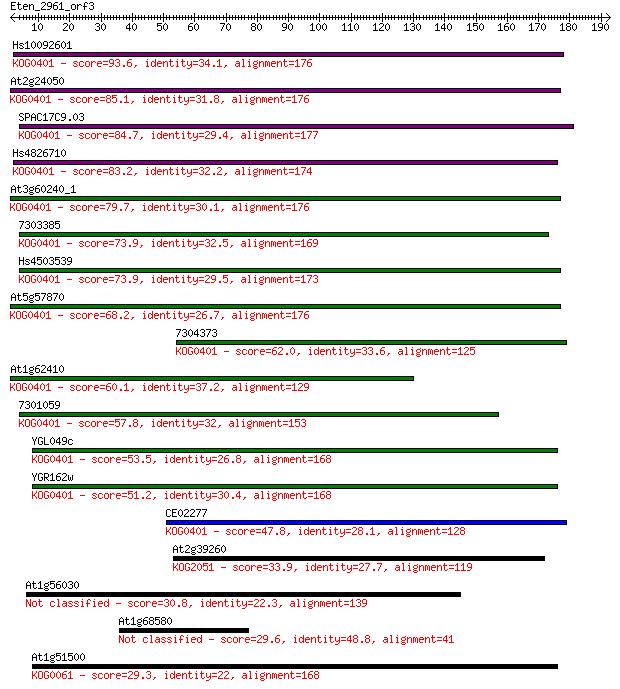
Sequences producing significant alignments: (Bits) Value
Hs10092601 93.6 2e-19
At2g24050 85.1 7e-17
SPAC17C9.03 84.7 1e-16
Hs4826710 83.2 3e-16
At3g60240_1 79.7 3e-15
7303385 73.9 2e-13
Hs4503539 73.9 2e-13
At5g57870 68.2 1e-11
7304373 62.0 6e-10
At1g62410 60.1 3e-09
7301059 57.8 1e-08
YGL049c 53.5 2e-07
YGR162w 51.2 1e-06
CE02277 47.8 2e-05
At2g39260 33.9 0.20
At1g56030 30.8 1.7
At1g68580 29.6 3.4
At1g51500 29.3 5.5
> Hs10092601
Length=1585
Score = 93.6 bits (231), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 60/186 (32%), Positives = 102/186 (54%), Gaps = 19/186 (10%)
Query 2 INFHRTFVNRCQDEFERLQGKNVLL----------ITEEERAECQDADDEAKLLKKKKTR 51
+NF + +NRCQ EFE+ + + + EER D +EAK K + R
Sbjct 831 VNFRKLLLNRCQKEFEKDKADDDVFEKKQKELEAASAPEERTRLHDELEEAK--DKARRR 888
Query 52 VLGNMRFIGELFLRRALSPNVLNDVVHALVFSSKGDAFPDEHFIECLTELLTTIGYTMDS 111
+GN++FIGELF + L+ +++D V L+ + DE +ECL LLTTIG +D
Sbjct 889 SIGNIKFIGELFKLKMLTEAIMHDCVVKLLKNH------DEESLECLCRLLTTIGKDLDF 942
Query 112 QDSSRAMMNEFIGKLQELQLRAGYSSRITFKIQDLLDLRQRHWTKKVFRDVAKSVAQIRE 171
+ ++ M+++ +++++ SSRI F +QD++DLR +W + K++ QI +
Sbjct 943 E-KAKPRMDQYFNQMEKIVKERKTSSRIRFMLQDVIDLRLCNWVSRRADQGPKTIEQIHK 1001
Query 172 DAKRDE 177
+AK +E
Sbjct 1002 EAKIEE 1007
> At2g24050
Length=747
Score = 85.1 bits (209), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 56/182 (30%), Positives = 93/182 (51%), Gaps = 16/182 (8%)
Query 1 KINFHRTFVNRCQDEFERLQGKNVLLITEEERAECQDADDEAKLLKKKKT---RVLGNMR 57
+I F R +N CQ+ FE G L +EE + + D E + + K+K R LGN+R
Sbjct 252 EITFKRVLLNNCQEAFE---GAGKL---KEEIRQMTNPDQEMERMDKEKMAKLRTLGNIR 305
Query 58 FIGELFLRRALSPNVLNDVVHALVFSSKGD---AFPDEHFIECLTELLTTIGYTMDSQDS 114
IGEL ++ + +++ +V L+ GD A P E +E L + TIG +D
Sbjct 306 LIGELLKQKMVPEKIVHHIVQELL----GDDTKACPAEGDVEALCQFFITIGKQLDDSPR 361
Query 115 SRAMMNEFIGKLQELQLRAGYSSRITFKIQDLLDLRQRHWTKKVFRDVAKSVAQIREDAK 174
SR + + + G+L+EL R+ F +Q+++DLR W + AK + +I +A+
Sbjct 362 SRGINDTYFGRLKELARHPQLELRLRFMVQNVVDLRANKWVPRREEVKAKKINEIHSEAE 421
Query 175 RD 176
R+
Sbjct 422 RN 423
> SPAC17C9.03
Length=1403
Score = 84.7 bits (208), Expect = 1e-16, Method: Composition-based stats.
Identities = 52/178 (29%), Positives = 92/178 (51%), Gaps = 7/178 (3%)
Query 4 FHRTFVNRCQDEFERLQGKNVLLITEEERAECQDADDEAKLLKKKKTRVLGNMRFIGELF 63
F + ++RCQ++FER N + + E + DE + K R LG +RFIGELF
Sbjct 1102 FRKYLLSRCQEDFERGWKAN---LPSGKAGEAEIMSDEYYVAAAIKRRGLGLVRFIGELF 1158
Query 64 LRRALSPNVLNDVVHALVFSSKGDAFPDEHFIECLTELLTTIGYTMDSQDSSRAMMNEFI 123
LS ++++ + L+ + P+E IE L LL T+G +D+ + A M+ ++
Sbjct 1159 KLSMLSEKIMHECIKRLLGNVTD---PEEEEIESLCRLLMTVGVNIDATEKGHAAMDVYV 1215
Query 124 GKLQELQLRAGYSSRITFKIQDLLDLRQRHW-TKKVFRDVAKSVAQIREDAKRDELMG 180
+++ + SRI F + D++D R+ W K K++A+I E+A+R + +
Sbjct 1216 LRMETITKIPNLPSRIKFMLMDVMDSRKNGWAVKNEVEKGPKTIAEIHEEAERKKALA 1273
> Hs4826710
Length=1396
Score = 83.2 bits (204), Expect = 3e-16, Method: Composition-based stats.
Identities = 56/190 (29%), Positives = 104/190 (54%), Gaps = 27/190 (14%)
Query 2 INFHRTFVNRCQDEFERLQGKNVLLITEEERAECQDA------------DDEAKLLKKKK 49
+NF + +NRCQ EFE+ + + + E+++ E +A +EA+ + ++
Sbjct 641 VNFRKLLLNRCQKEFEKDKDDDEVF--EKKQKEMDEAATAEERERLKEELEEARDIARRC 698
Query 50 TRVLGNMRFIGELFLRRALSPNVLNDVVHALVFSSKGDAFPDEHFIECLTELLTTIGYTM 109
+ LGN++FIGELF + L+ +++D V L+ + DE +ECL LLTTIG +
Sbjct 699 S--LGNIKFIGELFKLKMLTEAIMHDCVVKLLKNH------DEESLECLCRLLTTIGKDL 750
Query 110 DSQDSSRAMMNEFIGKLQELQLRAGYSSRITFKIQDLLDLRQRHWTKKVF----RDVAKS 165
D + ++ M+++ +++++ SSRI F +QD+LDLR +W + R + +S
Sbjct 751 DFE-KAKPRMDQYFNQMEKIIKEKKTSSRIRFMLQDVLDLRGSNWVPRRGDQGPRPLTRS 809
Query 166 VAQIREDAKR 175
+ ++R R
Sbjct 810 IRRLRWKTSR 819
> At3g60240_1
Length=1528
Score = 79.7 bits (195), Expect = 3e-15, Method: Composition-based stats.
Identities = 53/178 (29%), Positives = 103/178 (57%), Gaps = 7/178 (3%)
Query 1 KINFHRTFVNRCQDEFERLQGKNVLL--ITEEERAECQDADDEAKLLKKKKTRVLGNMRF 58
KI F R +N+CQ+EFER + + + EE + E + + E K L+ ++ R+LGN+R
Sbjct 987 KITFKRLLLNKCQEEFERGEKEEEEASRVAEEGQVEQTEEEREEKRLQVRR-RMLGNIRL 1045
Query 59 IGELFLRRALSPNVLNDVVHALVFSSKGDAFPDEHFIECLTELLTTIGYTMDSQDSSRAM 118
IGEL+ +R L+ +++ + L+ ++ P E IE L +L++TIG +D + ++
Sbjct 1046 IGELYKKRMLTEKIMHACIQKLLGYNQD---PHEENIEALCKLMSTIGVMID-HNKAKFQ 1101
Query 119 MNEFIGKLQELQLRAGYSSRITFKIQDLLDLRQRHWTKKVFRDVAKSVAQIREDAKRD 176
M+ + K++ L + SSR+ F + + +DLR+ W +++ + K + ++ DA ++
Sbjct 1102 MDGYFEKMKMLSCKQELSSRVRFMLINAIDLRKNKWQERMKVEGPKKIEEVHRDAAQE 1159
> 7303385
Length=869
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 55/175 (31%), Positives = 85/175 (48%), Gaps = 12/175 (6%)
Query 4 FHRTFVNRCQDEFERLQGKNVLLITEEE-RAECQDADDEAKLLKKKKTRVLGNMRFIGEL 62
F R + C+D+F N L E + R ++ DE + K R+LGN++FIGEL
Sbjct 26 FLRLLIAVCRDKF-----NNRLKRDENDNRPPPENEADEEERRHLAKQRMLGNVKFIGEL 80
Query 63 FLRRALSPNVLNDVVHALVFSSKGDAFPDEHF---IECLTELLTTIGYTMDSQDSSRAMM 119
LS NVL+ + L K + +ECL +LL T G +DS+ + +M
Sbjct 81 NKLDMLSKNVLHQCIMELFDKKKKRTAGTQEMCEDMECLAQLLKTCGKNLDSE-QGKELM 139
Query 120 NEFIGKLQELQLRAGYSSRITFKIQDLLDLRQRHWTKKVFRDVAKSVA--QIRED 172
N++ KL+ + Y RI F ++D+++LRQ +W + V QIR D
Sbjct 140 NQYFEKLERRSKSSEYPPRIRFMLKDVIELRQNNWVPRKLGTTEGPVPIKQIRSD 194
> Hs4503539
Length=907
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 51/176 (28%), Positives = 99/176 (56%), Gaps = 8/176 (4%)
Query 4 FHRTFVNRCQDEFERLQGKNVLLITEEERAECQDADDEAKLLKKKKTRVLGNMRFIGELF 63
F R +++ QDEFE + +NV + + E + +++ + K ++LGN++FIGEL
Sbjct 163 FRRLLISKLQDEFEN-RTRNVDVYDKRENPLLPEEEEQRAI---AKIKMLGNIKFIGELG 218
Query 64 LRRALSPNVLNDVVHALVFSSKGDAFPD-EHFIECLTELLTTIGYTMDSQDSSRAMMNEF 122
+ ++L+ + L+ K D +ECL +++ T+G +D + ++++M+++
Sbjct 219 KLDLIHESILHKCIKTLLEKKKRVQLKDMGEDLECLCQIMRTVGPRLD-HERAKSLMDQY 277
Query 123 IGKLQELQLRAGYSSRITFKIQDLLDLRQRHW-TKKVFRD-VAKSVAQIREDAKRD 176
++ L L +RI F +QD ++LR+ HW +K F D K++ QIR+DA +D
Sbjct 278 FARMCSLMLSKELPARIRFLLQDTVELREHHWVPRKAFLDNGPKTINQIRQDAVKD 333
> At5g57870
Length=780
Score = 68.2 bits (165), Expect = 1e-11, Method: Composition-based stats.
Identities = 47/179 (26%), Positives = 88/179 (49%), Gaps = 10/179 (5%)
Query 1 KINFHRTFVNRCQDEFERLQGKNVLLITEEERAECQDADDEAK---LLKKKKTRVLGNMR 57
+I F R +N CQ+ FE G + L EE + D EA+ K K + LGN+R
Sbjct 293 EITFKRVLLNICQEAFE---GASQL---REELRQMSAPDQEAERNDKEKLLKLKTLGNIR 346
Query 58 FIGELFLRRALSPNVLNDVVHALVFSSKGDAFPDEHFIECLTELLTTIGYTMDSQDSSRA 117
IGEL ++ + +++ +V L+ + + P E +E + TIG +D S+
Sbjct 347 LIGELLKQKMVPEKIVHHIVQELLGADE-KVCPAEENVEAICHFFKTIGKQLDGNVKSKR 405
Query 118 MMNEFIGKLQELQLRAGYSSRITFKIQDLLDLRQRHWTKKVFRDVAKSVAQIREDAKRD 176
+ + + +LQ L R+ F +Q+++D+R W + A+++ +I +A+++
Sbjct 406 INDVYFKRLQALSKNPQLELRLRFMVQNIIDMRSNGWVPRREEMKARTITEIHTEAEKN 464
> 7304373
Length=1666
Score = 62.0 bits (149), Expect = 6e-10, Method: Composition-based stats.
Identities = 42/136 (30%), Positives = 73/136 (53%), Gaps = 18/136 (13%)
Query 54 GNMRFIGELFLRRALSPNVLNDVVHALVFSSKGDAFPDEHFIECLTELLTTIGYT----- 108
G +RFIGELF L+ ++ + L+ E +ECL +LLTT+G
Sbjct 895 GTVRFIGELFKISMLTGKIIYSCIDTLLNPHS------EDMLECLCKLLTTVGAKFEKTP 948
Query 109 MDSQDSSRAM-MNEFIGKLQELQLR-----AGYSSRITFKIQDLLDLRQRHWTKKVFRDV 162
++S+D SR + + I ++Q + + A SSR+ F +QD++DLR+ W + +
Sbjct 949 VNSKDPSRCYSLEKSITRMQAIASKTDKDGARVSSRVRFMLQDVIDLRKNKW-QTSRNEA 1007
Query 163 AKSVAQIREDAKRDEL 178
K++ QI ++AK ++L
Sbjct 1008 PKTMGQIEKEAKNEQL 1023
> At1g62410
Length=223
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 48/134 (35%), Positives = 72/134 (53%), Gaps = 16/134 (11%)
Query 1 KINFHRTFVNRCQDEFERLQGKNVLLITEEER---AECQDAD--DEAKLLKKKKTRVLGN 55
+I+F R +N CQ FER ++EE R A Q+A+ DE +LL R LGN
Sbjct 54 EISFKRVLLNTCQKVFERTDD-----LSEEIRKMNAPDQEAEREDEVRLLN---LRTLGN 105
Query 56 MRFIGELFLRRALSPNVLNDVVHALVFSSKGDAFPDEHFIECLTELLTTIGYTMDSQDSS 115
+RF GELFL+R L+ V+ + L+ ++ P E I + L T+G +DS +S
Sbjct 106 LRFCGELFLKRMLTEKVVLAIGQKLLEDAE-QMCPSEEKIIAICLFLNTVGKKLDSLNSK 164
Query 116 RAMMNEFIGKLQEL 129
+MNE + +L+ L
Sbjct 165 --LMNEILRRLKNL 176
> 7301059
Length=1905
Score = 57.8 bits (138), Expect = 1e-08, Method: Composition-based stats.
Identities = 49/180 (27%), Positives = 82/180 (45%), Gaps = 33/180 (18%)
Query 4 FHRTFVNRCQDEFE-RLQGKNV----LLITEEERAECQDADDEAKLLKKK-------KTR 51
F + + R Q EFE + N L T E +C D +A+L + + R
Sbjct 1180 FTSSLITRIQHEFESNVNDANAKSKKLQPTMERINQCSDPAKKAELRAEMEDLEYQFRRR 1239
Query 52 VLGNMRFIGELFLRRALSPNVLNDVVHALVFSSKGDAFPDEHFIECLTELLTTIGYTMDS 111
G +RFIGELF ++L+ + + + V +L+ E +E + +LLTT+G+ ++S
Sbjct 1240 AWGTVRFIGELFKLQSLTNDRVLNCVESLLEHGC------EEKLEYMCKLLTTVGHLLES 1293
Query 112 QDSSRAMMNEFIGKL---------------QELQLRAGYSSRITFKIQDLLDLRQRHWTK 156
+ + I K+ Q+ SSR+ F +QD+LDLR R+W +
Sbjct 1294 SLPEHYQLRDRIEKIFRRIQDIINRSRGTSHRQQVHIKISSRVRFMMQDVLDLRLRNWDQ 1353
> YGL049c
Length=914
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 45/177 (25%), Positives = 78/177 (44%), Gaps = 11/177 (6%)
Query 8 FVNRCQDEFERLQGKNVLLITEEERAECQDADDEAKLLKKKKTRVLGNMRFIGELFLRRA 67
V RC +EFE+ + + E + DE + K R LG +RFIG L+
Sbjct 659 LVARCHEEFEKGWADKLPAGEDGNPLEPEMMSDEYYIAAAAKRRGLGLVRFIGYLYCLNL 718
Query 68 LSPNVLNDVVHALVFSSKGDAFPDEHFIECLTELLTTIG--------YTMDSQDSSRAMM 119
L+ ++ + L+ D P E +E + ELL T+G T + ++
Sbjct 719 LTGKMMFECFRRLMKDLNND--PSEETLESVIELLNTVGEQFEHDKFVTPQATLEGSVLL 776
Query 120 NEFIGKLQELQLRAGYSSRITFKIQDLLDLRQ-RHWTKKVFRDVAKSVAQIREDAKR 175
+ LQ + S+RI FK+ D+ +LR+ +HW K++ QI ++ ++
Sbjct 777 DNLFMLLQHIIDGGTISNRIKFKLIDVKELREIKHWNSAKKDAGPKTIQQIHQEEEQ 833
> YGR162w
Length=952
Score = 51.2 bits (121), Expect = 1e-06, Method: Composition-based stats.
Identities = 51/179 (28%), Positives = 83/179 (46%), Gaps = 15/179 (8%)
Query 8 FVNRCQDEFERLQGKNVLLITEEERA--ECQDADDEAKLLKKKKTRVLGNMRFIGELFLR 65
V RC EF++ G L T E+ E + +E K R LG +RFIG L+
Sbjct 699 LVARCHAEFDK--GWTDKLPTNEDGTPLEPEMMSEEYYAAASAKRRGLGLVRFIGFLYRL 756
Query 66 RALSPNVLNDVVHALVFSSKGDAFPDEHFIECLTELLTTIG--YTMDSQDSSRA------ 117
L+ ++ + L+ P E +E + ELL T+G + DS + +A
Sbjct 757 NLLTGKMMFECFRRLMKDLTDS--PSEETLESVVELLNTVGEQFETDSFRTGQATLEGSQ 814
Query 118 MMNEFIGKLQELQLRAGYSSRITFKIQDLLDLRQ-RHWTKKVFRDVAKSVAQIREDAKR 175
+++ G L + A SSRI FK+ D+ +LR ++W + K++ QI E+ +R
Sbjct 815 LLDSLFGILDNIIQTAKISSRIKFKLIDIKELRHDKNWNSDKKDNGPKTIQQIHEEEER 873
> CE02277
Length=1156
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 36/134 (26%), Positives = 68/134 (50%), Gaps = 8/134 (5%)
Query 51 RVLGNMRFIGELFLRRALSPNVLNDVVHALVFSSKGDAFP----DEHFIECLTELLTTIG 106
R G M F+G L+ + LS ++ L S K DE I C +L+ T+G
Sbjct 643 RKFGVMTFMGYLYRNQLLSTKIVQTCTFELFNSIKDQDIKKEDVDEESIHCGLQLIETVG 702
Query 107 YTMD-SQDSSRAMMNEFIGKLQELQLRAGYSSRITFKIQDLLDLRQRHW-TKKVFRDVAK 164
+D S+DS+ ++++ KL+ + S++I F I +L++LR+ W +K K
Sbjct 703 VMLDKSKDSTTVFLDQWFQKLE--AAKPYCSNKIRFMIMNLMELRKDKWIPRKSTESGPK 760
Query 165 SVAQIREDAKRDEL 178
+ +I +D +++++
Sbjct 761 KIDEIHKDIRQEKI 774
> At2g39260
Length=1029
Score = 33.9 bits (76), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 33/128 (25%), Positives = 62/128 (48%), Gaps = 10/128 (7%)
Query 53 LGNMRFIGELFLRRALSPNVLNDVVHALVF----SSKGDAF-PDEHF--IECLTELLTTI 105
L +MRF+GEL+ + +V+ + ++ + +S+ + P E F + + LL T
Sbjct 608 LAHMRFLGELYNYEHVDSSVIFETLYLTLLYGHDTSEQEVLDPPEDFFRVRMVIILLETC 667
Query 106 GYTMDSQDSSRAMMNEFIGKLQELQLRAGY-SSRITFKIQDLL-DLRQRHWTKKVFRDVA 163
G+ D + SS+ +++F+ Q L G+ I F +QDL +LR +V
Sbjct 668 GHYFD-RGSSKKRLDQFLIHFQRYILSKGHLPLDIEFDLQDLFANLRPNMTRYSTIDEVN 726
Query 164 KSVAQIRE 171
++ Q+ E
Sbjct 727 AAILQLEE 734
> At1g56030
Length=371
Score = 30.8 bits (68), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 31/139 (22%), Positives = 52/139 (37%), Gaps = 18/139 (12%)
Query 6 RTFVNRCQDEFERLQGKNVLLITEEERAECQDADDEAKLLKKKKTRVLGNMRFIGELFLR 65
R F+ CQ E E L K L + DE+K R + +R + E+F
Sbjct 33 RQFLRNCQMELENLSLKTELPLV---------YVDESKYW-----RFIKTVRVLAEVFNN 78
Query 66 RALSPNVLNDVVHALVFSSKGDAFPDEHFIECLTELLTTIGYTMDSQDSSRAMMNEFIGK 125
+ ++ L+ P E + + L+TI D Q S F+
Sbjct 79 MKTTRTTRKSIIQVLM----NPILPSERSTDAMNLFLSTIEKLADLQFSDEDFNQLFVSS 134
Query 126 LQELQLRAGYSSRITFKIQ 144
+LQL Y+ ++ K++
Sbjct 135 RLDLQLENKYNDKVEVKLR 153
> At1g68580
Length=704
Score = 29.6 bits (65), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 28/48 (58%), Gaps = 7/48 (14%)
Query 36 QDADDEAKLLKKK--KTRV-----LGNMRFIGELFLRRALSPNVLNDV 76
QDADDE+K L++ +RV LG++R G +R L P+ NDV
Sbjct 403 QDADDESKKLEEWILTSRVAAGDHLGDLRIKGRKVVRPMLKPSKENDV 450
> At1g51500
Length=687
Score = 29.3 bits (64), Expect = 5.5, Method: Composition-based stats.
Identities = 37/179 (20%), Positives = 63/179 (35%), Gaps = 25/179 (13%)
Query 8 FVNRCQDEFERLQGKNVLLITEEERAECQDADDEAKLLKKKKTRVLGNMRFIGELFLRRA 67
FV + R G+ V+ + +E D+ LL +T G +F E F
Sbjct 205 FVIQALRNIARDGGRTVVSSIHQPSSEVFALFDDLFLLSSGETVYFGESKFAVEFFAEAG 264
Query 68 LSPNVLNDVVHALVFSSKGDAFPDEHFIECLTELLTTIGYTM-------DSQDSSRAMMN 120
F P +HF+ C+ T+ T+ ++ +S +MN
Sbjct 265 --------------FPCPKKRNPSDHFLRCINSDFDTVTATLKGSQRIRETPATSDPLMN 310
Query 121 ----EFIGKLQELQLRAGYSSRITFKIQDLLDLRQRHWTKKVFRDVAKSVAQIREDAKR 175
E +L E R+ Y+ +I++L + H + A Q+R KR
Sbjct 311 LATSEIKARLVENYRRSVYAKSAKSRIRELASIEGHHGMEVRKGSEATWFKQLRTLTKR 369
Lambda K H
0.322 0.137 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3238438140
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40