bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2966_orf2
Length=159
Score E
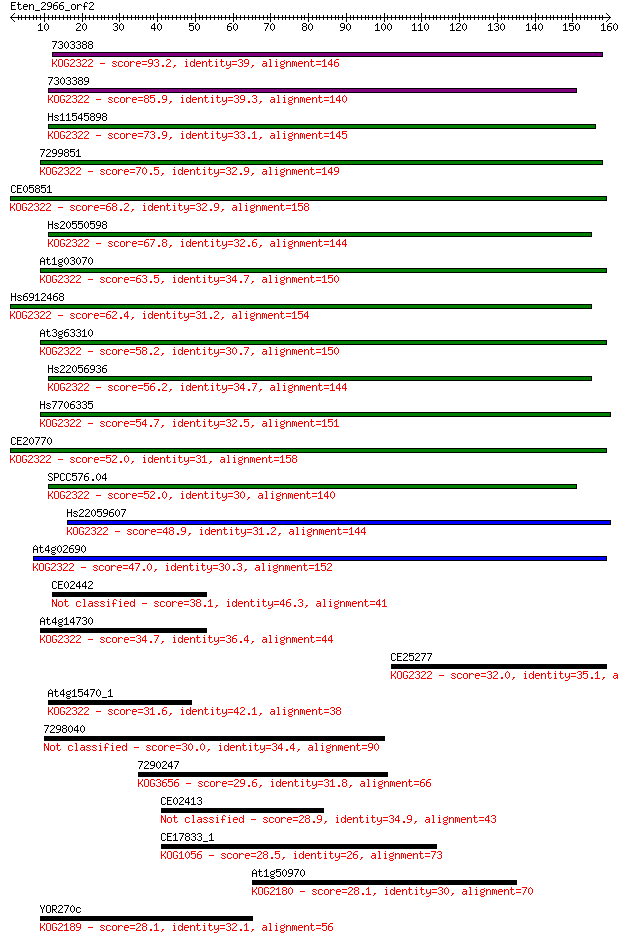
Sequences producing significant alignments: (Bits) Value
7303388 93.2 2e-19
7303389 85.9 3e-17
Hs11545898 73.9 1e-13
7299851 70.5 1e-12
CE05851 68.2 7e-12
Hs20550598 67.8 8e-12
At1g03070 63.5 2e-10
Hs6912468 62.4 3e-10
At3g63310 58.2 7e-09
Hs22056936 56.2 2e-08
Hs7706335 54.7 8e-08
CE20770 52.0 5e-07
SPCC576.04 52.0 5e-07
Hs22059607 48.9 4e-06
At4g02690 47.0 1e-05
CE02442 38.1 0.007
At4g14730 34.7 0.071
CE25277 32.0 0.55
At4g15470_1 31.6 0.64
7298040 30.0 1.8
7290247 29.6 2.7
CE02413 28.9 4.6
CE17833_1 28.5 5.5
At1g50970 28.1 7.5
YOR270c 28.1 7.8
> 7303388
Length=324
Score = 93.2 bits (230), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 57/146 (39%), Positives = 81/146 (55%), Gaps = 0/146 (0%)
Query 12 IRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQNYWLAIVAAVCGLVLQLVLVC 71
IR F+RKV IL Q++ TFG ++F + +TF N WL VA LV L + C
Sbjct 109 IRRGFIRKVYLILMGQLIVTFGAVALFVYHEGTKTFARNNMWLFWVALGVMLVTMLSMAC 168
Query 72 VPDLARKVPVNFILMTLITACYAVLTSCAAAASSWESFLIAIGSTFVVVVGLMLFACQTK 131
+ R+ P NFI + L TA + L +A + + L+A+G T V + L +FA QTK
Sbjct 169 CESVRRQTPTNFIFLGLFTAAQSFLMGVSATKYAPKEVLMAVGITAAVCLALTIFALQTK 228
Query 132 YDFTGCGTYLFVAVLCLMIFGILSIF 157
YDFT G L ++ +IFGI++IF
Sbjct 229 YDFTMMGGILIACMVVFLIFGIVAIF 254
> 7303389
Length=239
Score = 85.9 bits (211), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 55/143 (38%), Positives = 77/143 (53%), Gaps = 9/143 (6%)
Query 11 QIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQN---YWLAIVAAVCGLVLQL 67
IR F+RKV IL Q+L TFG SVF F + ++ +N +W+A+ + V +
Sbjct 23 SIRKGFIRKVYLILMCQLLITFGFVSVFTFSKASQEWVQKNPALFWIALAVLI---VTMI 79
Query 68 VLVCVPDLARKVPVNFILMTLITACYAVLTSCAAAASSWESFLIAIGSTFVVVVGLMLFA 127
+ C + RK P+NFI + L T + L A + L+A+G T V +GL LFA
Sbjct 80 CMACCESVRRKTPLNFIFLFLFTVAESFLLGMVAGQFEADEVLMAVGITAAVALGLTLFA 139
Query 128 CQTKYDFTGCGTYLFVAVLCLMI 150
QTKYDFT CG V V CL++
Sbjct 140 LQTKYDFTMCGG---VLVACLVV 159
> Hs11545898
Length=311
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 48/145 (33%), Positives = 80/145 (55%), Gaps = 0/145 (0%)
Query 11 QIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQNYWLAIVAAVCGLVLQLVLV 70
++RH F+RKV I++VQ+L T I ++F FV + F+ +N + V+ +V L+L
Sbjct 93 KVRHTFIRKVYSIISVQLLITVAIIAIFTFVEPVSAFVRRNVAVYYVSYAVFVVTYLILA 152
Query 71 CVPDLARKVPVNFILMTLITACYAVLTSCAAAASSWESFLIAIGSTFVVVVGLMLFACQT 130
C R+ P N IL+TL T +T ++ ++ +IA+ T VV + + +F QT
Sbjct 153 CCQGPRRRFPWNIILLTLFTFAMGFMTGTISSMYQTKAVIIAMIITAVVSISVTIFCFQT 212
Query 131 KYDFTGCGTYLFVAVLCLMIFGILS 155
K DFT C V + L++ GI++
Sbjct 213 KVDFTSCTGLFCVLGIVLLVTGIVT 237
> 7299851
Length=264
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 49/149 (32%), Positives = 77/149 (51%), Gaps = 0/149 (0%)
Query 9 SVQIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQNYWLAIVAAVCGLVLQLV 68
S IR F+RKV IL Q++ + + +R + ++ W+ +VA + + +
Sbjct 47 SASIRRGFIRKVYLILLAQLITSLVVIVSLTADKRVRLMVAESTWIFVVAILIVVFSLVA 106
Query 69 LVCVPDLARKVPVNFILMTLITACYAVLTSCAAAASSWESFLIAIGSTFVVVVGLMLFAC 128
L C DL R+ P NFI ++ T + L AA + +A+ T V +GL LFA
Sbjct 107 LGCNEDLRRQTPANFIFLSAFTIAESFLLGVAACRYAPMEIFMAVLITASVCLGLTLFAL 166
Query 129 QTKYDFTGCGTYLFVAVLCLMIFGILSIF 157
QT+YDFT G L ++ L+ FGI++IF
Sbjct 167 QTRYDFTVMGGLLVSCLIILLFFGIVTIF 195
> CE05851
Length=244
Score = 68.2 bits (165), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 52/163 (31%), Positives = 84/163 (51%), Gaps = 9/163 (5%)
Query 1 DTKITDHISVQ-IRHAFVRKVLGILAVQILFTFGIASVFGFVP----TLRTFLLQNYWLA 55
D K H S Q +R AFVRKV ++ + F I + F +P + ++ N+W+
Sbjct 15 DGKYNLHFSSQTVRAAFVRKVFMLVTIM----FAITAAFCVIPMVSEPFQDWVKNNFWVY 70
Query 56 IVAAVCGLVLQLVLVCVPDLARKVPVNFILMTLITACYAVLTSCAAAASSWESFLIAIGS 115
+A + LV+ + L C +L R+ PVN IL+T+ T AV+T A + +S LI +
Sbjct 71 FIAIIVFLVVAIALSCCGNLRRQFPVNIILLTIFTLSAAVMTMFVTACYNVQSVLICLCI 130
Query 116 TFVVVVGLMLFACQTKYDFTGCGTYLFVAVLCLMIFGILSIFF 158
T V +++F+ +TK D T F+ + L FGI ++ F
Sbjct 131 TTVCSGSVIIFSMKTKSDLTSKMGIAFMLSMVLFSFGIFALIF 173
> Hs20550598
Length=316
Score = 67.8 bits (164), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 47/147 (31%), Positives = 76/147 (51%), Gaps = 6/147 (4%)
Query 11 QIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQN---YWLAIVAAVCGLVLQL 67
++R FVRKV IL +Q+L T + ++F F ++ ++ N YW + L
Sbjct 97 KVRRVFVRKVYTILLIQLLVTLAVVALFTFCDPVKDYVQANPGWYWASYAVF---FATYL 153
Query 68 VLVCVPDLARKVPVNFILMTLITACYAVLTSCAAAASSWESFLIAIGSTFVVVVGLMLFA 127
L C R P N IL+T+ T A LT ++ + S L+ +G T +V + + +F+
Sbjct 154 TLACCSGPRRHFPWNLILLTVFTLSMAYLTGMLSSYYNTTSVLLCLGITALVCLSVTVFS 213
Query 128 CQTKYDFTGCGTYLFVAVLCLMIFGIL 154
QTK+DFT C LFV ++ L G++
Sbjct 214 FQTKFDFTSCQGVLFVLLMTLFFSGLI 240
> At1g03070
Length=247
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 52/161 (32%), Positives = 79/161 (49%), Gaps = 19/161 (11%)
Query 9 SVQIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQNYWLAIVAAVCGLVLQLV 68
S ++R F+RKV I+A Q+L T +AS FV + F GL L +V
Sbjct 31 SPELRWGFIRKVYSIIAFQLLATIAVASTVVFVRPIAVFF--------ATTSAGLALWIV 82
Query 69 LVCVPDLA--------RKVPVNFILMTLITACYAVLTSCAAAASSWESFLIAIGSTFVVV 120
L+ P + +K PVN++L+ + T A A +S + L A T VVV
Sbjct 83 LIITPLIVMCPLYYYHQKHPVNYLLLGIFTVALAFAVGLTCAFTSGKVILEAAILTTVVV 142
Query 121 VGLMLF---ACQTKYDFTGCGTYLFVAVLCLMIFGILSIFF 158
+ L ++ A + YDF G +LF A++ LM+F ++ IFF
Sbjct 143 LSLTVYTFWAAKKGYDFNFLGPFLFGALIVLMVFALIQIFF 183
> Hs6912468
Length=316
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 48/155 (30%), Positives = 78/155 (50%), Gaps = 2/155 (1%)
Query 1 DTKITDHISVQIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLL-QNYWLAIVAA 59
+ ++ ++ ++R FVRKV IL +Q+L T + ++F F + Q W A
Sbjct 87 SSPLSAGMTKKVRRVFVRKVYTILLIQLLVTLAVVALFTFCDPCQGLCSGQPGWYWASYA 146
Query 60 VCGLVLQLVLVCVPDLARKVPVNFILMTLITACYAVLTSCAAAASSWESFLIAIGSTFVV 119
V L L C R P N IL+T+ T A LT ++ + S L+ +G T +V
Sbjct 147 VF-FATYLTLACCSGPRRHFPWNLILLTVFTLSMAYLTGMLSSYYNTTSVLLCLGITALV 205
Query 120 VVGLMLFACQTKYDFTGCGTYLFVAVLCLMIFGIL 154
+ + +F+ QTK+DFT C LFV ++ L G++
Sbjct 206 CLSVTVFSFQTKFDFTSCQGVLFVLLMTLFFSGLI 240
> At3g63310
Length=239
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 46/161 (28%), Positives = 79/161 (49%), Gaps = 19/161 (11%)
Query 9 SVQIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQNYWLAIVAAVCGLVLQLV 68
S ++R +F+RKV I+++Q+L T +A+ V ++ F G L ++
Sbjct 22 SPELRWSFIRKVYSIISIQLLVTIAVAATVVKVHSISVFF--------TTTTAGFALYIL 73
Query 69 LVCVPDLA--------RKVPVNFILMTLITACYAVLTSCAAAASSWESFLIAIGSTFVVV 120
L+ P + +K PVN++L+ + T A A +S + L ++ T VVV
Sbjct 74 LILTPLIVMCPLYYYHQKHPVNYLLLGIFTVALAFAVGLTCAFTSGKVILESVILTAVVV 133
Query 121 VGLMLF---ACQTKYDFTGCGTYLFVAVLCLMIFGILSIFF 158
+ L L+ A + +DF G +LF AV+ LM+F + I F
Sbjct 134 ISLTLYTFWAAKRGHDFNFLGPFLFGAVIVLMVFSFIQILF 174
> Hs22056936
Length=349
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 50/172 (29%), Positives = 78/172 (45%), Gaps = 29/172 (16%)
Query 11 QIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQN---YWLAIVAAVCGLVLQL 67
++R FVRKV IL +Q+L T + ++F F ++ ++ N YW A A L L
Sbjct 101 KVRRVFVRKVYTILLIQLLVTLAVVALFTFCDPVKDYVQANPGWYW-ASYAVFFATYLTL 159
Query 68 VLVCVPDLA-------------------------RKVPVNFILMTLITACYAVLTSCAAA 102
P A R P N IL+T+ T A LT ++
Sbjct 160 ACCSGPRYANGLRRNLWEYEQDHENSGPSPASHQRHFPWNLILLTVFTLSMAYLTGMLSS 219
Query 103 ASSWESFLIAIGSTFVVVVGLMLFACQTKYDFTGCGTYLFVAVLCLMIFGIL 154
+ S L+ +G T +V + + +F+ QTK+DFT C LFV ++ L G++
Sbjct 220 YYNTTSVLLCLGITALVCLSVTVFSFQTKFDFTSCQGVLFVLLMTLFFSGLI 271
> Hs7706335
Length=258
Score = 54.7 bits (130), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 49/155 (31%), Positives = 82/155 (52%), Gaps = 11/155 (7%)
Query 9 SVQIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQNYWLAIVAAV--CGLVLQ 66
+V IR AF+RKV IL++Q+L T ++VF + ++RTF+ ++ L ++ A+ GL+
Sbjct 47 TVHIRMAFLRKVYSILSLQVLLTTVTSTVFLYFESVRTFVHESPALILLFALGSLGLIFA 106
Query 67 LVLVCVPDLARKVPVNFILMTLITACYAVLTSCAAAASSWESFLI--AIGSTFVVVVGLM 124
L L K P+N L+ T A+ + A + ++ ++I A T V GL
Sbjct 107 LTLN-----RHKYPLNLYLLFGFTLLEAL--TVAVVVTFYDVYIILQAFILTTTVFFGLT 159
Query 125 LFACQTKYDFTGCGTYLFVAVLCLMIFGILSIFFH 159
++ Q+K DF+ G LF + L + G L FF+
Sbjct 160 VYTLQSKKDFSKFGAGLFALLWILCLSGFLKFFFY 194
> CE20770
Length=295
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 49/167 (29%), Positives = 78/167 (46%), Gaps = 13/167 (7%)
Query 1 DTKITDHISVQ-IRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQNYWLAIVAA 59
D K + S + +R AFVRKV ++ + + + + + +N L + +
Sbjct 67 DGKYSFQFSDKTVRAAFVRKVFSLVFIMLCIVAAVTVIPWVHDDTMRMVRRNSALYLGSY 126
Query 60 VCGLVLQLVLVCVPDLARKVPVNFILMTLITACYAVLTSCAAAASSWESFLIA----IGS 115
V V L LVC + RK PVN I+ + T +V+T +A L+A IG
Sbjct 127 VIFFVTYLSLVCCEGVRRKFPVNLIVTGIFTLATSVMTMVISAHHDANVVLLALAICIGC 186
Query 116 TFVVVVGLMLFACQTKYDFTGCGTYLFVAVLCLMIFGIL----SIFF 158
TF +V+ A QTK+D T Y+ + +C M FG++ S+FF
Sbjct 187 TFSIVI----VASQTKFDLTAHMGYILIISMCFMFFGLVVVICSMFF 229
> SPCC576.04
Length=266
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 42/140 (30%), Positives = 64/140 (45%), Gaps = 3/140 (2%)
Query 11 QIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQNYWLAIVAAVCGLVLQLVLV 70
IR AF+RKV IL Q+ T +F P ++ + W I+ LV+ L+
Sbjct 53 SIRMAFLRKVYAILTAQLFVTSLFGGIFYLHPAFSFWVQMHPWFLILNFFISLVVLFGLI 112
Query 71 CVPDLARKVPVNFILMTLITACYAVLTSCAAAASSWESFLIAIGSTFVVVVGLMLFACQT 130
P P N+I + L TA + A S L A+ T V V L F Q+
Sbjct 113 MKP---YSYPRNYIFLFLFTALEGLTLGTAITFFSARIILEAVFITLGVFVALTAFTFQS 169
Query 131 KYDFTGCGTYLFVAVLCLMI 150
K+DF+ G +L+V++ L++
Sbjct 170 KWDFSRLGGFLYVSLWSLIL 189
> Hs22059607
Length=285
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 45/148 (30%), Positives = 78/148 (52%), Gaps = 11/148 (7%)
Query 16 FVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQNYWLAIVAAV--CGLVLQLVLVCVP 73
F+RKV IL++Q+L T ++VF + ++RTF+ ++ L ++ A+ GL+ L+L
Sbjct 81 FLRKVYSILSLQVLLTTVTSTVFLYFESVRTFVHESPALILLFALGSLGLIFALILN--- 137
Query 74 DLARKVPVNFILMTLITACYAVLTSCAAAASSWESFLI--AIGSTFVVVVGLMLFACQTK 131
K P+N L+ T A+ + A + ++ ++I A T V GL ++ Q+K
Sbjct 138 --RHKYPLNLYLLFGFTLLEAL--TVAVVVTFYDVYIILQAFILTTTVFFGLTVYTLQSK 193
Query 132 YDFTGCGTYLFVAVLCLMIFGILSIFFH 159
DF+ G LF + L + G L FF+
Sbjct 194 KDFSKFGAGLFALLWILCLSGFLKFFFY 221
> At4g02690
Length=248
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 46/163 (28%), Positives = 78/163 (47%), Gaps = 19/163 (11%)
Query 7 HISVQIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQNYWLAIVAAVCGLVLQ 66
H + ++R F+RKV I+A Q+L T +A+ V + F GL L
Sbjct 29 HENPELRWGFIRKVYSIIAFQLLATVAVAATVVTVRPIALFF--------ATTGLGLALY 80
Query 67 LVLVCVPDLA--------RKVPVNFILMTLITACYAVLTSCAAAASSWESFLIAIGSTFV 118
+V++ P + +K PVN++L+ + T A + A ++ + L ++ T V
Sbjct 81 IVIIITPLIVLCPLYYYHQKHPVNYLLLGIFTLALAFVVGLTCAFTNGKVILESVILTSV 140
Query 119 VVVGLMLF---ACQTKYDFTGCGTYLFVAVLCLMIFGILSIFF 158
VV+ L L+ A + YDF G +LF A+ L+ F ++ I F
Sbjct 141 VVLSLTLYTFWAARKGYDFNFLGPFLFGALTVLIFFALIQILF 183
> CE02442
Length=130
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 19/41 (46%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 12 IRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQNY 52
IR AF+RKVLGI+ Q+LFT GI + +P L + Y
Sbjct 69 IRIAFLRKVLGIVGFQLLFTIGICAAIYNIPNSNQLLQKQY 109
> At4g14730
Length=100
Score = 34.7 bits (78), Expect = 0.071, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 30/44 (68%), Gaps = 0/44 (0%)
Query 9 SVQIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQNY 52
S ++R AF+RK+ IL++Q+L T G+++V FV + F+ + +
Sbjct 21 SSELRWAFIRKLYSILSLQLLVTVGVSAVVYFVRPIPEFITETH 64
> CE25277
Length=133
Score = 32.0 bits (71), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 32/57 (56%), Gaps = 0/57 (0%)
Query 102 AASSWESFLIAIGSTFVVVVGLMLFACQTKYDFTGCGTYLFVAVLCLMIFGILSIFF 158
A S ++ LI++ T ++LFA TK D T C F+ +CLM+FG+++ F
Sbjct 7 ATYSADAVLISLLITTGCSASIILFAATTKKDLTSCLGVAFILGICLMLFGLMACIF 63
> At4g15470_1
Length=331
Score = 31.6 bits (70), Expect = 0.64, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 11 QIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFL 48
Q+R F+RKV GIL+ Q+L T I++V P + L
Sbjct 44 QLRWGFIRKVYGILSAQLLLTTLISAVVVLNPPVNDLL 81
> 7298040
Length=529
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 31/94 (32%), Positives = 46/94 (48%), Gaps = 10/94 (10%)
Query 10 VQIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQNYWLAIVAAVCGLVLQLVL 69
V++R + + GI A+ I+ G+ + G R +LL +VA C V+ LV
Sbjct 10 VRLRLKVAQWIYGIAALFIVLAIGLLILPGL---FRRYLLVP---DVVATYCFFVIGLVT 63
Query 70 VCV----PDLARKVPVNFILMTLITACYAVLTSC 99
+CV L RK P N+I+ I AC A+ T C
Sbjct 64 LCVYVNVTWLRRKFPFNWIVSCCIAACLALGTVC 97
> 7290247
Length=634
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 21/66 (31%), Positives = 30/66 (45%), Gaps = 4/66 (6%)
Query 35 ASVFGFVPTLRTFLLQNYWLAIVAAVCGLVLQLVLVCVPDLARKVPVNFILMTLITACYA 94
A GF +L TF +L ++ +CG +L +V + R V FI I+ C A
Sbjct 26 ADATGFSQSLLTFAAVMTFLIMIVGICGNLLTVVALLKCPKVRNVAAAFI----ISLCIA 81
Query 95 VLTSCA 100
L CA
Sbjct 82 DLLFCA 87
> CE02413
Length=448
Score = 28.9 bits (63), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 26/49 (53%), Gaps = 6/49 (12%)
Query 41 VPTLRTFLLQNYWLA------IVAAVCGLVLQLVLVCVPDLARKVPVNF 83
+ +L+T L+ Y L+ +V C L L + ++C P+L R V +F
Sbjct 378 IESLQTRFLEEYMLSADLSSVLVVTACALRLPIYMLCNPELRRAVKKSF 426
> CE17833_1
Length=1130
Score = 28.5 bits (62), Expect = 5.5, Method: Composition-based stats.
Identities = 19/81 (23%), Positives = 39/81 (48%), Gaps = 8/81 (9%)
Query 41 VPTLRTFLLQNYWLAIVAAVCGLVLQLVLVCVPDLARKVPV--------NFILMTLITAC 92
+P + ++ + LA+V AV G++ + + V PV ++I+++ + AC
Sbjct 567 IPEVVSWTSFGHILALVLAVTGIITSMATLAVFLRHNSTPVVKSTTRELSYIILSGLVAC 626
Query 93 YAVLTSCAAAASSWESFLIAI 113
YAV + A S+ F+ +
Sbjct 627 YAVSFALLATPSTTSCFITRV 647
> At1g50970
Length=520
Score = 28.1 bits (61), Expect = 7.5, Method: Composition-based stats.
Identities = 21/72 (29%), Positives = 36/72 (50%), Gaps = 4/72 (5%)
Query 65 LQLVLVCVPDLARKVPVNFILMTLITACYAVLTSCAAAASSWESFLIAIGSTFVVVVGL- 123
++++L+ VP LAR+ + +L+ TA Y L + + E+ L I S V V+
Sbjct 355 MKMILLKVPSLARQPEASALLVKTATASYVKLVNHQMKRA--EAVLKVIASPIVTVIDTY 412
Query 124 -MLFACQTKYDF 134
LF +T +F
Sbjct 413 RALFPEETPMEF 424
> YOR270c
Length=840
Score = 28.1 bits (61), Expect = 7.8, Method: Composition-based stats.
Identities = 18/56 (32%), Positives = 26/56 (46%), Gaps = 5/56 (8%)
Query 9 SVQIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQNYWLAIVAAVCGLV 64
++ + HA + VL + +QI F F GFV T L W A+ AV L+
Sbjct 738 ALSLAHAQLSSVLWTMTIQIAFGFR-----GFVGVFMTVALFAMWFALTCAVLVLM 788
Lambda K H
0.337 0.146 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2136300674
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40