bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
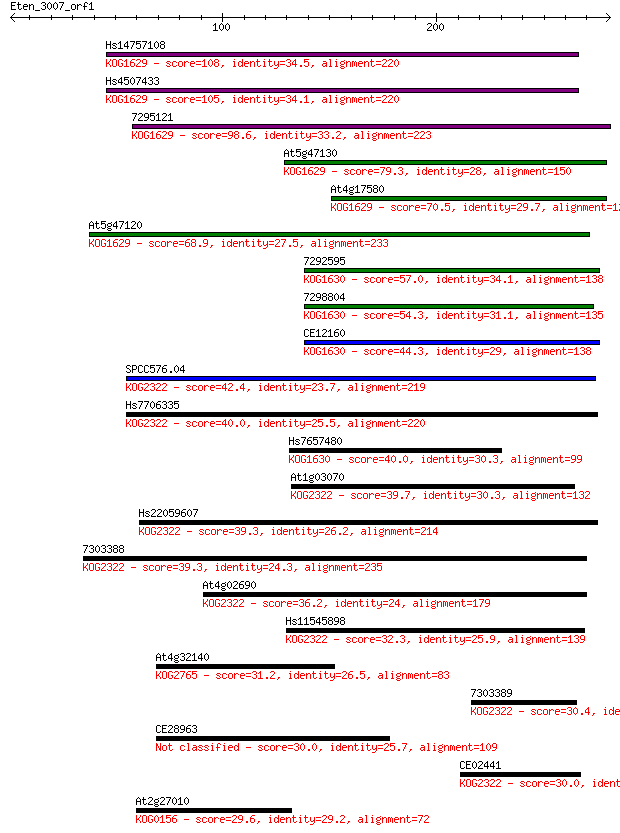
Query= Eten_3007_orf1
Length=280
Score E
Sequences producing significant alignments: (Bits) Value
Hs14757108 108 1e-23
Hs4507433 105 1e-22
7295121 98.6 1e-20
At5g47130 79.3 9e-15
At4g17580 70.5 4e-12
At5g47120 68.9 1e-11
7292595 57.0 5e-08
7298804 54.3 3e-07
CE12160 44.3 3e-04
SPCC576.04 42.4 0.001
Hs7706335 40.0 0.005
Hs7657480 40.0 0.005
At1g03070 39.7 0.006
Hs22059607 39.3 0.008
7303388 39.3 0.010
At4g02690 36.2 0.083
Hs11545898 32.3 1.2
At4g32140 31.2 2.7
7303389 30.4 3.9
CE28963 30.0 5.1
CE02441 30.0 6.3
At2g27010 29.6 7.8
> Hs14757108
Length=237
Score = 108 bits (271), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 76/221 (34%), Positives = 123/221 (55%), Gaps = 7/221 (3%)
Query 46 SLLDFSPLSNLQKKHLAKIYAALTSNVLATALGVYVHL-NCFALPNFLALLLSLACVLAL 104
+LL FS ++ ++HL K+YA+ + A G YVH+ F L+ L SL ++ L
Sbjct 12 ALLKFSHITPSTQQHLKKVYASFALCMFVAAAGAYVHMVTHFIQAGLLSALGSLILMIWL 71
Query 105 SFSSQKAHAESKMLTGERALYFGGFGFLNGMLAGNYLAAVHFYVGPQVVPAAFLASVAIF 164
+ E K R GF FL G+ G L V P ++P AF+ + IF
Sbjct 72 MATPHSHETEQK-----RLGLLAGFAFLTGVGLGPALEFC-IAVNPSILPTAFMGTAMIF 125
Query 165 CCLSAAALVAKQRSYLYLGSILGAAITYLSLASLANIFLRLQLLNDVLLWGGLFMYLGFV 224
C + +AL A++RSYL+LG IL +A++ L L+SL N+F L L+ GL + GFV
Sbjct 126 TCFTLSALYARRRSYLFLGGILMSALSLLLLSSLGNVFFGSIWLFQANLYVGLVVMCGFV 185
Query 225 VYDTQLAVAQFDAGNRDYLLQAVQFYINFISIFLRLVAILS 265
++DTQL + + + G++DY+ + +++FI++F +L+ IL+
Sbjct 186 LFDTQLIIEKAEHGDQDYIWHCIDLFLDFITVFRKLMMILA 226
> Hs4507433
Length=237
Score = 105 bits (261), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 75/221 (33%), Positives = 121/221 (54%), Gaps = 7/221 (3%)
Query 46 SLLDFSPLSNLQKKHLAKIYAALTSNVLATALGVYVHL-NCFALPNFLALLLSLACVLAL 104
+LL FS ++ ++HL K+YA+ + A G YVH+ F L+ L SL ++ L
Sbjct 12 ALLKFSHITPSTQQHLKKVYASFALCMFVAAAGAYVHMVTHFIQAGLLSALGSLILMIWL 71
Query 105 SFSSQKAHAESKMLTGERALYFGGFGFLNGMLAGNYLAAVHFYVGPQVVPAAFLASVAIF 164
+ E K R GF FL G+ G L V P ++P AF+ + IF
Sbjct 72 MATPHSHETEQK-----RLGLLAGFAFLTGVGLGPALEFC-IAVNPSILPTAFMGTAMIF 125
Query 165 CCLSAAALVAKQRSYLYLGSILGAAITYLSLASLANIFLRLQLLNDVLLWGGLFMYLGFV 224
C + +AL A++RSYL+LG IL +A++ L L+SL N+F L+ GL + GFV
Sbjct 126 TCFTLSALYARRRSYLFLGGILMSALSLLLLSSLGNVFFGSIWPFQANLYVGLVVMCGFV 185
Query 225 VYDTQLAVAQFDAGNRDYLLQAVQFYINFISIFLRLVAILS 265
+ DTQL + + + G++DY+ + +++FI++F +L+ IL+
Sbjct 186 LVDTQLIIEKAEHGDQDYIWHCIDLFLDFITVFRKLMMILA 226
> 7295121
Length=245
Score = 98.6 bits (244), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 74/223 (33%), Positives = 117/223 (52%), Gaps = 10/223 (4%)
Query 58 KKHLAKIYAALTSNVLATALGVYVHLNCFALPNFLALLLSLACVLALSFSSQKAHAESKM 117
++HL+K+Y L S ATA+G + + F LA + +L VL L F
Sbjct 28 REHLSKVYMVLGSTAAATAMGAMLQMRDFLDLGVLAAVATLVLVLGLHFYKDDGKNYYTR 87
Query 118 LTGERALYFGGFGFLNGMLAGNYLAAVHFYVGPQVVPAAFLASVAIFCCLSAAALVAKQR 177
L LY FGF +G G L + + P ++ +A + F LS +AL+A+Q
Sbjct 88 LG---MLY--AFGFCSGQTLGPLLGYIC-SINPAIILSALTGTFVTFISLSLSALLAEQG 141
Query 178 SYLYLGSILGAAITYLSLASLANIFLRLQLLNDVLLWGGLFMYLGFVVYDTQLAVAQFDA 237
YLYLG +L + I ++L SL N+ + + L+ G+F+ F+VYDTQ V +
Sbjct 142 KYLYLGGMLVSVINTMALLSLFNMVFKSYFVQVTQLYVGVFVMAAFIVYDTQNIVEKCRN 201
Query 238 GNRDYLLQAVQFYINFISIFLRLVAILSERQEETNRRKREERR 280
GNRD + A+ + + +S+F RL+ IL++++E RK+ ERR
Sbjct 202 GNRDVVQHALDLFFDVLSMFRRLLIILTQKEE----RKQNERR 240
> At5g47130
Length=187
Score = 79.3 bits (194), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 42/150 (28%), Positives = 84/150 (56%), Gaps = 3/150 (2%)
Query 129 FGFLNGMLAGNYLAAVHFYVGPQVVPAAFLASVAIFCCLSAAALVAKQRSYLYLGSILGA 188
FG L+G G + + + ++ AFL + IF C SA A++A++R Y+YLG +L +
Sbjct 40 FGVLHGASVGPCIKST-IDIDSSILITAFLGTAVIFFCFSAVAMLARRREYIYLGGLLSS 98
Query 189 AITYLSLASLANIFLRLQLLNDVLLWGGLFMYLGFVVYDTQLAVAQFDAGNRDYLLQAVQ 248
+ L+ ++ F + ++ ++ GL +++G +V +TQ + + G+ DY + ++
Sbjct 99 GFSLLTWLKNSDQFASATV--EIQMYLGLLLFVGCIVVNTQEIIEKAHCGDMDYAVHSLI 156
Query 249 FYINFISIFLRLVAILSERQEETNRRKREE 278
YI F+ +FL++++I+ + RR EE
Sbjct 157 LYIGFVRVFLQILSIMWNTSADRIRRNNEE 186
> At4g17580
Length=247
Score = 70.5 bits (171), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 38/129 (29%), Positives = 72/129 (55%), Gaps = 1/129 (0%)
Query 151 QVVPAAFLASVAIFCCLSAAALVAKQRSYLYLGSILGAAITYLSLASLAN-IFLRLQLLN 209
V+ AF+ + F C SAAA++A +R YLY G+ L ++ L +A+ IF +
Sbjct 119 SVLVTAFVGTAVAFVCFSAAAMLATRREYLYHGASLACCMSILWWVQIASSIFGGSTTVV 178
Query 210 DVLLWGGLFMYLGFVVYDTQLAVAQFDAGNRDYLLQAVQFYINFISIFLRLVAILSERQE 269
L+ GL +++G++V DTQ+ + G+ DY+ + F+ +F S+F++++ + R+
Sbjct 179 KFELYFGLLIFVGYIVVDTQMITEKAHHGDMDYVQHSFTFFTDFASLFVQILVLNMFRKM 238
Query 270 ETNRRKREE 278
+ R+ R
Sbjct 239 KKGRKDRRN 247
> At5g47120
Length=247
Score = 68.9 bits (167), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 64/240 (26%), Positives = 125/240 (52%), Gaps = 22/240 (9%)
Query 38 GAAAVPARSLLDFSPLSNLQKKHLAKIYAALTSNVLATALGVYVHLNCFALPNFLALLLS 97
G+ + SL +F +S + HL ++Y L ++A+A G Y+H+ L N +L +
Sbjct 13 GSRSWSYDSLKNFRQISPAVQNHLKRVYLTLCCALVASAFGAYLHV----LWNIGGILTT 68
Query 98 LACVLALSFS-SQKAHAESKMLTGERALYFGGFGFLNGMLAG---NYLAAVHFYVGPQVV 153
+ C+ + + S + K L+ F++ +L G L V V P ++
Sbjct 69 IGCIGTMIWLLSCPPYEHQKRLS---------LLFVSAVLEGASVGPLIKVAIDVDPSIL 119
Query 154 PAAFLASVAIFCCLSAAALVAKQRSYLYLGSILGAAIT---YLSLASLANIFLRLQLLND 210
AF+ + F C SAAA++A++R YLYLG +L + ++ +L AS +IF +
Sbjct 120 ITAFVGTAIAFVCFSAAAMLARRREYLYLGGLLSSGLSMLMWLQFAS--SIFGGSASIFK 177
Query 211 VLLWGGLFMYLGFVVYDTQLAVAQFDAGNRDYLLQAVQFYINFISIFLRLVAILSERQEE 270
L+ GL +++G++V DTQ + + G+ DY+ ++ + +F+++F+R++ I+ + +
Sbjct 178 FELYFGLLIFVGYMVVDTQEIIEKAHLGDMDYVKHSLTLFTDFVAVFVRILIIMLKNSAD 237
> 7292595
Length=341
Score = 57.0 bits (136), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 47/149 (31%), Positives = 75/149 (50%), Gaps = 16/149 (10%)
Query 138 GNYLAAVHFYVGPQVVPAAFLASVAIFCCLSAAALVAKQRSYLYLGSILGAAITYLSLAS 197
G +A + F GP + AA L + I LS A A +LY+G L + + +S
Sbjct 198 GAVIAPICFMGGPILTRAA-LYTGGIVGGLSTIAACAPSDKFLYMGGPLAIGLGVVFASS 256
Query 198 LANIFLRLQL-----LNDVLLWGGLFMYLGFVVYDTQLAV------AQFDAGNRDYLLQA 246
LA+++L L + L+GGL ++ GF++YDTQ V Q+ D + +
Sbjct 257 LASMWLPPTTALGAGLASMSLYGGLVLFSGFLLYDTQRMVRRAEVYPQYSYTPYDPINAS 316
Query 247 VQFYINFISIFLRLVAILSERQEETNRRK 275
+ Y++ ++IF+R+V ILS Q RRK
Sbjct 317 MSIYMDVLNIFIRIVTILSGGQ----RRK 341
> 7298804
Length=305
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 42/146 (28%), Positives = 72/146 (49%), Gaps = 12/146 (8%)
Query 138 GNYLAAVHFYVGPQVVPAAFLASVAIFCCLSAAALVAKQRSYLYLGSILGAAITYLSLAS 197
G LA + GP ++ A L + I LS A A +L++G L + + +S
Sbjct 161 GAVLAPMCLLGGP-ILTKALLYTSGIVGALSTVAACAPSEKFLHMGGPLAIGLGVVFASS 219
Query 198 LANIFLRLQL-----LNDVLLWGGLFMYLGFVVYDTQLAV------AQFDAGNRDYLLQA 246
LA+++L L + L+GGL ++ GF++YDTQ V Q+ D + A
Sbjct 220 LASMWLPPTTAVGAGLASMSLYGGLILFSGFLLYDTQRIVKSAELYPQYSKFPYDPINHA 279
Query 247 VQFYINFISIFLRLVAILSERQEETN 272
+ Y++ ++IF+R+ IL+ Q+ N
Sbjct 280 LAIYMDALNIFIRIAIILAGDQKRKN 305
> CE12160
Length=342
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 40/157 (25%), Positives = 69/157 (43%), Gaps = 22/157 (14%)
Query 138 GNYLAAVHFYVGPQVVPAAFLASVAIFCCLSAAALVAKQRSYLYLGSILGAAITYLSLAS 197
G A + F GP + AA+ + I LSA A+ A +L + L + +A+
Sbjct 189 GAVFAPLCFMAGPVLTRAAWY-TAGIVGGLSATAITAPSEKFLMMSGPLAMGFGVVFVAN 247
Query 198 LANIFLRL-----QLLNDVLLWGGLFMYLGFVVYDTQLAVAQFDAGNR------------ 240
+ FL L ++++GGL ++ F++YDTQ V + +
Sbjct 248 IGAFFLPPGSALGASLASIVVYGGLILFSAFLLYDTQRLVKKAENHPHSSQLYGSDMQIR 307
Query 241 --DYLLQAVQFYINFISIFLRLVAILSERQEETNRRK 275
D + + Y++ ++IF+RLV I+ NRRK
Sbjct 308 SFDPINAQMSIYMDVLNIFMRLVMIMGGMNG--NRRK 342
> SPCC576.04
Length=266
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 52/229 (22%), Positives = 107/229 (46%), Gaps = 25/229 (10%)
Query 55 NLQKKHLAKIYAALTSNVLATAL--GVYVHLNCFAL-----PNFLAL--LLSLACVLALS 105
+++ L K+YA LT+ + T+L G++ F+ P FL L +SL + L
Sbjct 53 SIRMAFLRKVYAILTAQLFVTSLFGGIFYLHPAFSFWVQMHPWFLILNFFISLVVLFGLI 112
Query 106 FSSQKAHAESKMLTGERALYFGGFGFLNGMLAGNYLAAVHFYVGPQVVPAAFLASVAIFC 165
K ++ + ++ F L G+ G A+ F+ ++ A F+ ++ +F
Sbjct 113 ---MKPYSYPR-----NYIFLFLFTALEGLTLG---TAITFFSARIILEAVFI-TLGVFV 160
Query 166 CLSAAALVAKQRSYLYLGSILGAAITYLSLASLANIFLRLQLLNDVLLWG-GLFMYLGFV 224
L+A +K + LG L ++ L L L F+ D+ G G ++ G++
Sbjct 161 ALTAFTFQSKW-DFSRLGGFLYVSLWSLILTPLIFFFVPSTPFIDMAFAGFGTLVFCGYI 219
Query 225 VYDTQLAVAQFDAGNRDYLLQAVQFYINFISIFLRLVAILSERQEETNR 273
++DT + ++ ++++ ++ Y++FI++F+R++ IL Q N
Sbjct 220 LFDTYNILHRYSP--EEFIMSSLMLYLDFINLFIRILQILGMLQNNDNN 266
> Hs7706335
Length=258
Score = 40.0 bits (92), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 56/229 (24%), Positives = 105/229 (45%), Gaps = 28/229 (12%)
Query 55 NLQKKHLAKIYAALTSNVLATALGVYVHLNCFALPNFL----ALLL-----SLACVLALS 105
+++ L K+Y+ L+ VL T + V L ++ F+ AL+L SL + AL+
Sbjct 49 HIRMAFLRKVYSILSLQVLLTTVTSTVFLYFESVRTFVHESPALILLFALGSLGLIFALT 108
Query 106 FSSQKAHAESKMLTGERALYFGGFGFLNGMLAGNYLAAVHFYVGPQVVPAAFLASVAIFC 165
+ K +L FGF +L +A V + ++ AF+ + +F
Sbjct 109 LNRHKYPLNLYLL----------FGF--TLLEALTVAVVVTFYDVYIILQAFILTTTVFF 156
Query 166 CLSAAALVAKQRSYLYLGSILGAAITYLSLASLANIFLRLQLLNDVLLWGGLFMYLGFVV 225
L+ L +K + + G+ L A + L L+ F +++ VL G ++ GF++
Sbjct 157 GLTVYTLQSK-KDFSKFGAGLFALLWILCLSGFLKFFFYSEIMELVLAAAGALLFCGFII 215
Query 226 YDTQLAVAQFDAGNRDYLLQAVQFYINFISIFLRLVAILSERQEETNRR 274
YDT + + +Y+L A+ Y++ I++FL L+ L E N++
Sbjct 216 YDTHSLMHKLSP--EEYVLAAISLYLDIINLFLHLLRFL----EAVNKK 258
> Hs7657480
Length=319
Score = 40.0 bits (92), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 30/104 (28%), Positives = 54/104 (51%), Gaps = 6/104 (5%)
Query 131 FLNGMLAGNYLAAVHFYVGPQVVPAAFLASVAIFCCLSAAALVAKQRSYLYLGSILGAAI 190
L+ + G +A + GP ++ AA+ + I LS A+ A +L +G+ LG +
Sbjct 194 LLHSGVMGAVVAPLTILGGPLLIRAAWY-TAGIVGGLSTVAMCAPSEKFLNMGAPLGVGL 252
Query 191 TYLSLASLANIFLRLQL-----LNDVLLWGGLFMYLGFVVYDTQ 229
+ ++SL ++FL L V ++GGL ++ F++YDTQ
Sbjct 253 GLVFVSSLGSMFLPPTTVAGATLYSVAMYGGLVLFSMFLLYDTQ 296
> At1g03070
Length=247
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 40/144 (27%), Positives = 74/144 (51%), Gaps = 16/144 (11%)
Query 132 LNGMLAGNYLAAVHFYVG--------PQVVPAAFLASVAIFCCLSAAALVAKQRSYLY-- 181
+N +L G + A+ F VG ++ AA L +V + L+ A ++ Y +
Sbjct 103 VNYLLLGIFTVALAFAVGLTCAFTSGKVILEAAILTTVVVLS-LTVYTFWAAKKGYDFNF 161
Query 182 LGSILGAAITYLSLASLANIFLRLQLLNDVLLWGGL--FMYLGFVVYDTQLAVAQFDAGN 239
LG L A+ L + +L IF L ++ V+++G L ++ G++VYDT + ++
Sbjct 162 LGPFLFGALIVLMVFALIQIFFPLGRIS-VMIYGCLAAIIFCGYIVYDTDNLIKRYS--Y 218
Query 240 RDYLLQAVQFYINFISIFLRLVAI 263
+Y+ AV Y++ I++FL L+ I
Sbjct 219 DEYIWAAVSLYLDIINLFLALLTI 242
> Hs22059607
Length=285
Score = 39.3 bits (90), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 56/223 (25%), Positives = 101/223 (45%), Gaps = 28/223 (12%)
Query 61 LAKIYAALTSNVLATALGVYVHLNCFALPNFL----ALLL-----SLACVLALSFSSQKA 111
L K+Y+ L+ VL T + V L ++ F+ AL+L SL + AL + K
Sbjct 82 LRKVYSILSLQVLLTTVTSTVFLYFESVRTFVHESPALILLFALGSLGLIFALILNRHKY 141
Query 112 HAESKMLTGERALYFGGFGFLNGMLAGNYLAAVHFYVGPQVVPAAFLASVAIFCCLSAAA 171
+L FGF +L +A V + ++ AF+ + +F L+
Sbjct 142 PLNLYLL----------FGF--TLLEALTVAVVVTFYDVYIILQAFILTTTVFFGLTVYT 189
Query 172 LVAKQRSYLYLGSILGAAITYLSLASLANIFLRLQLLNDVLLWGGLFMYLGFVVYDTQLA 231
L +K + + G+ L A + L L+ F +++ VL G ++ GF++YDT
Sbjct 190 LQSK-KDFSKFGAGLFALLWILCLSGFLKFFFYSEIMELVLAAAGALLFCGFIIYDTHSL 248
Query 232 VAQFDAGNRDYLLQAVQFYINFISIFLRLVAILSERQEETNRR 274
+ + +Y+L A+ Y++ I++FL L+ L E N++
Sbjct 249 MHKLSP--EEYVLAAISLYLDIINLFLHLLRFL----EAVNKK 285
> 7303388
Length=324
Score = 39.3 bits (90), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 57/249 (22%), Positives = 111/249 (44%), Gaps = 25/249 (10%)
Query 35 GVLGAAAVPARSLLDFS-PLSNLQKKHLAKIYAALTSNVLAT--ALGVYVH---LNCFAL 88
G GA P +FS ++++ + K+Y L ++ T A+ ++V+ FA
Sbjct 87 GGYGAYDDPESQPKNFSFDDQSIRRGFIRKVYLILMGQLIVTFGAVALFVYHEGTKTFAR 146
Query 89 PNFLALLLSLACVLALSFSSQKAHAES-KMLTGERALYFGGF----GFLNGMLAGNYLAA 143
N ++L +L S A ES + T ++ G F FL G+ A Y
Sbjct 147 NNMWLFWVALGVMLVTMLS--MACCESVRRQTPTNFIFLGLFTAAQSFLMGVSATKY--- 201
Query 144 VHFYVGPQVVPAAFLASVAIFCCLSAAALVAKQRSYLYLGSILGAAITYLSLASLANIFL 203
P+ V A + A+ L+ AL K + +G IL A + + + IF+
Sbjct 202 -----APKEVLMAVGITAAVCLALTIFALQTK-YDFTMMGGILIACMVVFLIFGIVAIFV 255
Query 204 RLQLLNDVLLWGGLFMYLGFVVYDTQLAVA---QFDAGNRDYLLQAVQFYINFISIFLRL 260
+ +++ V G ++ +++YDTQL + ++ +Y+ A+ Y++ I+IF+ +
Sbjct 256 KGKIITLVYASIGALLFSVYLIYDTQLMMGGEHKYSISPEEYIFAALNLYLDIINIFMYI 315
Query 261 VAILSERQE 269
+ I+ ++
Sbjct 316 LTIIGASRD 324
> At4g02690
Length=248
Score = 36.2 bits (82), Expect = 0.083, Method: Compositional matrix adjust.
Identities = 43/183 (23%), Positives = 84/183 (45%), Gaps = 18/183 (9%)
Query 91 FLALLLSLACVLALSFSSQKAHAESKMLTGERALYFGGFGFLNGMLAGNYLAAVHFYVGP 150
++ ++++ VL + + H + +L G L F+ G+ F G
Sbjct 80 YIVIIITPLIVLCPLYYYHQKHPVNYLLLGIFTL---ALAFVVGLTCA-------FTNGK 129
Query 151 QVVPAAFLASVAIFCCLSAAALVAKQRSYLY--LGSILGAAITYLSLASLANIFLRLQLL 208
++ + L SV + L+ A ++ Y + LG L A+T L +L I L +
Sbjct 130 VILESVILTSVVVLS-LTLYTFWAARKGYDFNFLGPFLFGALTVLIFFALIQILFPLGRV 188
Query 209 NDVLLWGGL--FMYLGFVVYDTQLAVAQFDAGNRDYLLQAVQFYINFISIFLRLVAILSE 266
+ V+++G L ++ G++VYDT + + +Y+ AV Y++ I++FL L+ +L
Sbjct 189 S-VMIYGCLVSIIFCGYIVYDTDNLIKRHTYD--EYIWAAVSLYLDIINLFLYLLTVLRA 245
Query 267 RQE 269
Q
Sbjct 246 LQR 248
> Hs11545898
Length=311
Score = 32.3 bits (72), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 36/146 (24%), Positives = 59/146 (40%), Gaps = 17/146 (11%)
Query 130 GFLNGMLAGNYLAAVHFYVGPQVVPAAFLASVAIFCCLSAAALVAKQRSYLYLGSILGAA 189
GF+ G ++ Y + ++ A SV IFC + + + LG +L
Sbjct 176 GFMTGTISSMYQTKA--VIIAMIITAVVSISVTIFCFQTKVDFTSCTGLFCVLGIVLLVT 233
Query 190 ITYLSLASLANIFLRLQLLNDVLLWGGLFMYLGFVVYDTQLAVAQFDAGNR-------DY 242
S+ L +L L G + F+ YDTQL + GNR DY
Sbjct 234 GIVTSIVLYFQYVYWLHMLYAAL---GAICFTLFLAYDTQLVL-----GNRKHTISPEDY 285
Query 243 LLQAVQFYINFISIFLRLVAILSERQ 268
+ A+Q Y + I IF ++ ++ +R
Sbjct 286 ITGALQIYTDIIYIFTFVLQLMGDRN 311
> At4g32140
Length=394
Score = 31.2 bits (69), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 22/86 (25%), Positives = 42/86 (48%), Gaps = 3/86 (3%)
Query 69 TSNVLATALGVYVHLNCFALPNFLALLLSLACVLALSFSSQKAHAESKM---LTGERALY 125
TS + +GV++ + L +A+ +S+A V+ + A ES++ L GER+L
Sbjct 180 TSGLFTLFIGVFLGQDTLNLSKVVAVFVSMAGVVMTTLGKTWASDESQLNSSLNGERSLM 239
Query 126 FGGFGFLNGMLAGNYLAAVHFYVGPQ 151
FG L+ + G + + + G +
Sbjct 240 GDLFGLLSAVSYGLFTVLLKKFAGEE 265
> 7303389
Length=239
Score = 30.4 bits (67), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 14/52 (26%), Positives = 31/52 (59%), Gaps = 3/52 (5%)
Query 216 GLFMYLGFVVYDTQLAVA---QFDAGNRDYLLQAVQFYINFISIFLRLVAIL 264
G ++ ++VYDTQL + ++ +Y+ A+ Y++ I+IF+ ++ I+
Sbjct 183 GALLFSVYLVYDTQLMLGGNHKYSISPEEYIFAALNLYLDIINIFMYILTII 234
> CE28963
Length=396
Score = 30.0 bits (66), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 28/110 (25%), Positives = 45/110 (40%), Gaps = 9/110 (8%)
Query 69 TSNVLATALGVYVHLNCFALPNFLALLLSLACVLALSFSSQKAHAESKMLTGERALYFGG 128
T+N +L VY H NC F+ +++ + L +F+ A K
Sbjct 33 TANCAPISLRVYPHFNC----RFVTMIVGILACLTATFTLGSVFAPIKRFPSSDGFMVQF 88
Query 129 FGFLNGMLAGNYLAAVHFYVG-PQVVPAAFLASVAIFCCLSAAALVAKQR 177
F L G + A+H +G P + P A L A++C +A A + R
Sbjct 89 FMSLGGFITS---LAIHALLGFPPIYPLAMLGG-ALWCAANAFAFLIMNR 134
> CE02441
Length=184
Score = 30.0 bits (66), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 17/75 (22%), Positives = 35/75 (46%), Gaps = 21/75 (28%)
Query 211 VLLWGGLF-------------------MYLGFVVYDTQLAVAQFDAGNRDYLLQAVQFYI 251
VLLW G+F ++ +V D + + +F DY+ V Y+
Sbjct 109 VLLWAGIFQMFFMSPAVNFVINVFGAGLFCVLLVIDLDMIMYRFSP--EDYICACVSLYM 166
Query 252 NFISIFLRLVAILSE 266
+ +++F+R++ I++E
Sbjct 167 DILNLFIRILQIVAE 181
> At2g27010
Length=498
Score = 29.6 bits (65), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 36/73 (49%), Gaps = 4/73 (5%)
Query 60 HLAKIYAALTSNVLATALGVYVHLNCFALPNFLALLLSLACVLALSFSSQKAHAESKML- 118
HL ++ + L++ G ++L+ F +P LL+S + F +Q + S+
Sbjct 50 HLLSLFMHRSLQKLSSKYGPLLYLHVFNVP---ILLVSSPSIAYEIFRTQDVNVSSRDFP 106
Query 119 TGERALYFGGFGF 131
T E +L FG FGF
Sbjct 107 TNEGSLLFGSFGF 119
Lambda K H
0.328 0.139 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6127321958
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40