bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
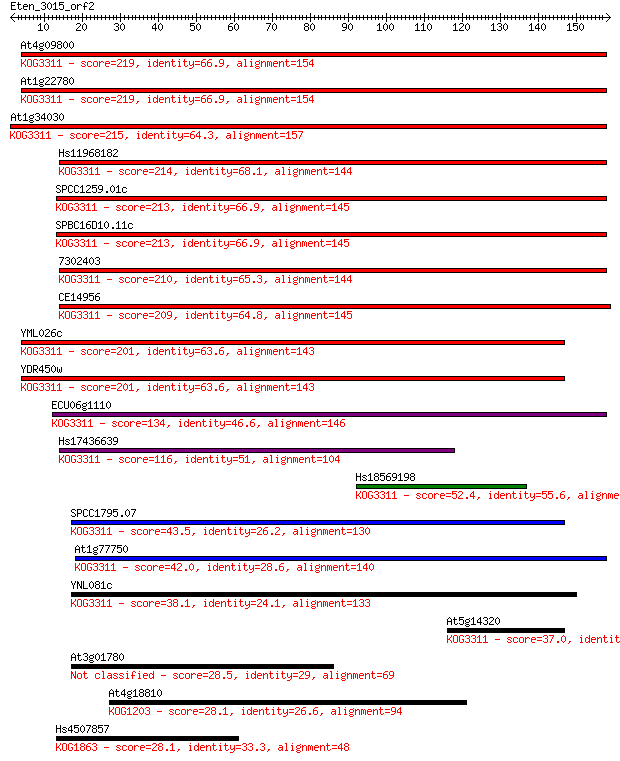
Query= Eten_3015_orf2
Length=158
Score E
Sequences producing significant alignments: (Bits) Value
At4g09800 219 2e-57
At1g22780 219 2e-57
At1g34030 215 2e-56
Hs11968182 214 8e-56
SPCC1259.01c 213 2e-55
SPBC16D10.11c 213 2e-55
7302403 210 8e-55
CE14956 209 3e-54
YML026c 201 3e-52
YDR450w 201 3e-52
ECU06g1110 134 5e-32
Hs17436639 116 2e-26
Hs18569198 52.4 4e-07
SPCC1795.07 43.5 2e-04
At1g77750 42.0 4e-04
YNL081c 38.1 0.006
At5g14320 37.0 0.014
At3g01780 28.5 5.2
At4g18810 28.1 7.6
Hs4507857 28.1 7.9
> At4g09800
Length=152
Score = 219 bits (558), Expect = 2e-57, Method: Compositional matrix adjust.
Identities = 103/154 (66%), Positives = 127/154 (82%), Gaps = 2/154 (1%)
Query 4 MSLRATNMADFQHILRVLNTNVDGREKVVIALTAIRGIGRRISTLICKKADIDTDKRAGE 63
MSL A +FQHILRVLNTNVDG++K++ ALT+I+GIGRR++ ++CKKAD+D +KRAGE
Sbjct 1 MSLVANE--EFQHILRVLNTNVDGKQKIMFALTSIKGIGRRLANIVCKKADVDMNKRAGE 58
Query 64 LTADEISKIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMR 123
L+A EI ++ IVA+P QF IP WFLNRQKD KDGK V +N LD R+D ERLKK+R
Sbjct 59 LSAAEIDNLMTIVANPRQFKIPDWFLNRQKDYKDGKYSQVVSNALDMKLRDDLERLKKIR 118
Query 124 LHRGLRHYWGLKVRGQHTKTTGRHGRTVGVAKKK 157
HRGLRHYWGL+VRGQHTKTTGR G+TVGV+KK+
Sbjct 119 NHRGLRHYWGLRVRGQHTKTTGRRGKTVGVSKKR 152
> At1g22780
Length=152
Score = 219 bits (558), Expect = 2e-57, Method: Compositional matrix adjust.
Identities = 103/154 (66%), Positives = 127/154 (82%), Gaps = 2/154 (1%)
Query 4 MSLRATNMADFQHILRVLNTNVDGREKVVIALTAIRGIGRRISTLICKKADIDTDKRAGE 63
MSL A +FQHILRVLNTNVDG++K++ ALT+I+GIGRR++ ++CKKAD+D +KRAGE
Sbjct 1 MSLVANE--EFQHILRVLNTNVDGKQKIMFALTSIKGIGRRLANIVCKKADVDMNKRAGE 58
Query 64 LTADEISKIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMR 123
L+A EI ++ IVA+P QF IP WFLNRQKD KDGK V +N LD R+D ERLKK+R
Sbjct 59 LSAAEIDNLMTIVANPRQFKIPDWFLNRQKDYKDGKYSQVVSNALDMKLRDDLERLKKIR 118
Query 124 LHRGLRHYWGLKVRGQHTKTTGRHGRTVGVAKKK 157
HRGLRHYWGL+VRGQHTKTTGR G+TVGV+KK+
Sbjct 119 NHRGLRHYWGLRVRGQHTKTTGRRGKTVGVSKKR 152
> At1g34030
Length=237
Score = 215 bits (548), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 101/157 (64%), Positives = 127/157 (80%), Gaps = 2/157 (1%)
Query 1 NSKMSLRATNMADFQHILRVLNTNVDGREKVVIALTAIRGIGRRISTLICKKADIDTDKR 60
+S MSL A +FQHILRVLNTNVDG++K++ ALT+I+GIGRR++ ++CKKAD+D +KR
Sbjct 11 SSVMSLVANE--EFQHILRVLNTNVDGKQKIMFALTSIKGIGRRLANIVCKKADVDMNKR 68
Query 61 AGELTADEISKIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLK 120
AGEL+A EI ++ IVA+P QF IP WFLNRQKD KDGK V +N LD R+D ERLK
Sbjct 69 AGELSAAEIDNLMTIVANPRQFKIPDWFLNRQKDYKDGKYSQVVSNALDMKLRDDLERLK 128
Query 121 KMRLHRGLRHYWGLKVRGQHTKTTGRHGRTVGVAKKK 157
K+R HRGLRHYWGL+VRGQHTKTTGR G+TV + +K+
Sbjct 129 KIRNHRGLRHYWGLRVRGQHTKTTGRRGKTVALDQKR 165
> Hs11968182
Length=152
Score = 214 bits (544), Expect = 8e-56, Method: Compositional matrix adjust.
Identities = 98/144 (68%), Positives = 119/144 (82%), Gaps = 0/144 (0%)
Query 14 FQHILRVLNTNVDGREKVVIALTAIRGIGRRISTLICKKADIDTDKRAGELTADEISKIV 73
FQHILRVLNTN+DGR K+ A+TAI+G+GRR + ++ +KADID KRAGELT DE+ +++
Sbjct 9 FQHILRVLNTNIDGRRKIAFAITAIKGVGRRYAHVVLRKADIDLTKRAGELTEDEVERVI 68
Query 74 AIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRHYWG 133
I+ +P Q+ IP WFLNRQKDVKDGK V AN LD+ RED ERLKK+R HRGLRH+WG
Sbjct 69 TIMQNPRQYKIPDWFLNRQKDVKDGKYSQVLANGLDNKLREDLERLKKIRAHRGLRHFWG 128
Query 134 LKVRGQHTKTTGRHGRTVGVAKKK 157
L+VRGQHTKTTGR GRTVGV+KKK
Sbjct 129 LRVRGQHTKTTGRRGRTVGVSKKK 152
> SPCC1259.01c
Length=152
Score = 213 bits (541), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 97/145 (66%), Positives = 121/145 (83%), Gaps = 0/145 (0%)
Query 13 DFQHILRVLNTNVDGREKVVIALTAIRGIGRRISTLICKKADIDTDKRAGELTADEISKI 72
+FQHILR+LNTNVDG+ KV+ A+T I+G+GRR + ++CKKADID KRAGELT +E+ +I
Sbjct 8 NFQHILRLLNTNVDGKVKVMFAMTQIKGVGRRYANIVCKKADIDMSKRAGELTTEELERI 67
Query 73 VAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRHYW 132
V I+ +P+QF IPSWFLNRQKD+ DGKS + ANN+DS RED ERLKK++ HRGLRH
Sbjct 68 VTIIQNPSQFKIPSWFLNRQKDINDGKSFQLLANNVDSKLREDLERLKKIQTHRGLRHAL 127
Query 133 GLKVRGQHTKTTGRHGRTVGVAKKK 157
L+VRGQHTKTTGR G+TVGV+KKK
Sbjct 128 DLRVRGQHTKTTGRRGKTVGVSKKK 152
> SPBC16D10.11c
Length=152
Score = 213 bits (541), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 97/145 (66%), Positives = 121/145 (83%), Gaps = 0/145 (0%)
Query 13 DFQHILRVLNTNVDGREKVVIALTAIRGIGRRISTLICKKADIDTDKRAGELTADEISKI 72
+FQHILR+LNTNVDG+ KV+ A+T I+G+GRR + ++CKKADID KRAGELT +E+ +I
Sbjct 8 NFQHILRLLNTNVDGKVKVMFAMTQIKGVGRRYANIVCKKADIDMSKRAGELTTEELERI 67
Query 73 VAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRHYW 132
V I+ +P+QF IPSWFLNRQKD+ DGKS + ANN+DS RED ERLKK++ HRGLRH
Sbjct 68 VTIIQNPSQFKIPSWFLNRQKDINDGKSFQLLANNVDSKLREDLERLKKIQTHRGLRHAL 127
Query 133 GLKVRGQHTKTTGRHGRTVGVAKKK 157
L+VRGQHTKTTGR G+TVGV+KKK
Sbjct 128 DLRVRGQHTKTTGRRGKTVGVSKKK 152
> 7302403
Length=152
Score = 210 bits (535), Expect = 8e-55, Method: Compositional matrix adjust.
Identities = 94/144 (65%), Positives = 122/144 (84%), Gaps = 0/144 (0%)
Query 14 FQHILRVLNTNVDGREKVVIALTAIRGIGRRISTLICKKADIDTDKRAGELTADEISKIV 73
FQHILR++NTN+DG+ KV IA+TAI+G+GRR S ++ KKAD+D KRAGE T +E+ K+V
Sbjct 9 FQHILRIMNTNIDGKRKVGIAMTAIKGVGRRYSNIVLKKADVDLTKRAGECTEEEVDKVV 68
Query 74 AIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRHYWG 133
I+++P Q+ +P+WFLNRQKD+ DGK ++++NLDS R+D ERLKK+R HRGLRHYWG
Sbjct 69 TIISNPLQYKVPNWFLNRQKDIIDGKYWQLTSSNLDSKLRDDLERLKKIRSHRGLRHYWG 128
Query 134 LKVRGQHTKTTGRHGRTVGVAKKK 157
L+VRGQHTKTTGR GRTVGV+KKK
Sbjct 129 LRVRGQHTKTTGRRGRTVGVSKKK 152
> CE14956
Length=154
Score = 209 bits (531), Expect = 3e-54, Method: Compositional matrix adjust.
Identities = 94/145 (64%), Positives = 120/145 (82%), Gaps = 0/145 (0%)
Query 14 FQHILRVLNTNVDGREKVVIALTAIRGIGRRISTLICKKADIDTDKRAGELTADEISKIV 73
FQHI RV+NTN+DG KV ALTAI+G+GRR + + C+KAD+D +KRAGELT ++ KIV
Sbjct 9 FQHIHRVMNTNIDGNRKVPYALTAIKGVGRRFAFVCCRKADVDVNKRAGELTEEDFDKIV 68
Query 74 AIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRHYWG 133
I+ +P+Q+ IP+WFLNRQKD+KDGK+ + + +D+ RED ER+KK+RLHRGLRHYWG
Sbjct 69 TIMQNPSQYKIPNWFLNRQKDIKDGKTGQLLSTAVDNKLREDLERMKKIRLHRGLRHYWG 128
Query 134 LKVRGQHTKTTGRHGRTVGVAKKKA 158
L+VRGQHTKTTGR GRTVGV+KKK
Sbjct 129 LRVRGQHTKTTGRKGRTVGVSKKKG 153
> YML026c
Length=146
Score = 201 bits (512), Expect = 3e-52, Method: Compositional matrix adjust.
Identities = 91/143 (63%), Positives = 112/143 (78%), Gaps = 0/143 (0%)
Query 4 MSLRATNMADFQHILRVLNTNVDGREKVVIALTAIRGIGRRISTLICKKADIDTDKRAGE 63
MSL FQHILR+LNTNVDG K+V ALT I+G+GRR S L+CKKAD+D KRAGE
Sbjct 1 MSLVVQEQGSFQHILRLLNTNVDGNIKIVYALTTIKGVGRRYSNLVCKKADVDLHKRAGE 60
Query 64 LTADEISKIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMR 123
LT +E+ +IV I+ +PT + IP+WFLNRQ D+ DGK H ANN++S R+D ERLKK+R
Sbjct 61 LTQEELERIVQIMQNPTHYKIPAWFLNRQNDITDGKDYHTLANNVESKLRDDLERLKKIR 120
Query 124 LHRGLRHYWGLKVRGQHTKTTGR 146
HRG+RH+WGL+VRGQHTKTTGR
Sbjct 121 AHRGIRHFWGLRVRGQHTKTTGR 143
> YDR450w
Length=146
Score = 201 bits (512), Expect = 3e-52, Method: Compositional matrix adjust.
Identities = 91/143 (63%), Positives = 112/143 (78%), Gaps = 0/143 (0%)
Query 4 MSLRATNMADFQHILRVLNTNVDGREKVVIALTAIRGIGRRISTLICKKADIDTDKRAGE 63
MSL FQHILR+LNTNVDG K+V ALT I+G+GRR S L+CKKAD+D KRAGE
Sbjct 1 MSLVVQEQGSFQHILRLLNTNVDGNIKIVYALTTIKGVGRRYSNLVCKKADVDLHKRAGE 60
Query 64 LTADEISKIVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMR 123
LT +E+ +IV I+ +PT + IP+WFLNRQ D+ DGK H ANN++S R+D ERLKK+R
Sbjct 61 LTQEELERIVQIMQNPTHYKIPAWFLNRQNDITDGKDYHTLANNVESKLRDDLERLKKIR 120
Query 124 LHRGLRHYWGLKVRGQHTKTTGR 146
HRG+RH+WGL+VRGQHTKTTGR
Sbjct 121 AHRGIRHFWGLRVRGQHTKTTGR 143
> ECU06g1110
Length=153
Score = 134 bits (338), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 68/146 (46%), Positives = 95/146 (65%), Gaps = 0/146 (0%)
Query 12 ADFQHILRVLNTNVDGREKVVIALTAIRGIGRRISTLICKKADIDTDKRAGELTADEISK 71
A+ Q+I+R+ NTN+DG +++ ALT IRGIG RI+T ICK+ ID KRAGE+ DE+ +
Sbjct 8 AEVQYIVRIHNTNIDGTKRIPFALTKIRGIGIRIATAICKRLGIDLRKRAGEMGEDEMKR 67
Query 72 IVAIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRHY 131
I + P IP ++N Q+D+ DG + H+ LD+ R ER KK + R R
Sbjct 68 ISDAILDPASVGIPESYMNHQRDIIDGTTSHLIGTRLDADLRMMIERGKKNKRIRAYRLD 127
Query 132 WGLKVRGQHTKTTGRHGRTVGVAKKK 157
GLKVRGQ TK+ GR GR++GV++KK
Sbjct 128 VGLKVRGQRTKSNGRRGRSMGVSRKK 153
> Hs17436639
Length=112
Score = 116 bits (291), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 53/104 (50%), Positives = 71/104 (68%), Gaps = 0/104 (0%)
Query 14 FQHILRVLNTNVDGREKVVIALTAIRGIGRRISTLICKKADIDTDKRAGELTADEISKIV 73
FQ ILRV TN+D K+ A+TAI+G+G+R + ++ +KADID R GELT DE+ ++
Sbjct 9 FQRILRVFKTNIDEWCKITFAITAIKGVGQRYAHVVLRKADIDLTSRVGELTEDEVECVI 68
Query 74 AIVASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFE 117
++ +P Q+ IP WFLNRQKDVKDGK AN LD+ ED E
Sbjct 69 TMMQNPHQYKIPGWFLNRQKDVKDGKYSQALANGLDNKLHEDLE 112
> Hs18569198
Length=139
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 25/45 (55%), Positives = 31/45 (68%), Gaps = 0/45 (0%)
Query 92 QKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRHYWGLKV 136
QKD K G V AN+LD+ RED E+LKK+ RGL H+WGL+V
Sbjct 95 QKDGKAGNYSQVLANDLDNKLREDLEQLKKIPACRGLHHFWGLRV 139
> SPCC1795.07
Length=151
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 34/130 (26%), Positives = 56/130 (43%), Gaps = 23/130 (17%)
Query 17 ILRVLNTNVDGREKVVIALTAIRGIGRRISTLICKKADIDTDKRAGELTADEISKIVAIV 76
++ +L V+ + V AL + GIG + IC K R ELT +++ + ++
Sbjct 1 MVYILGVAVNDDKPVRFALLSFYGIGHAKAEEICAKLSFHNTLRVRELTNVQLTTLSQLL 60
Query 77 ASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRHYWGLKV 136
+ T L RQ++ D RL +R +RG+RH GL V
Sbjct 61 SGMTI----EGDLRRQRNA-------------------DISRLVNIRCYRGMRHVNGLPV 97
Query 137 RGQHTKTTGR 146
GQ+T+T +
Sbjct 98 NGQNTRTNAK 107
> At1g77750
Length=154
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 40/142 (28%), Positives = 60/142 (42%), Gaps = 26/142 (18%)
Query 18 LRVLNTNVDGREKVVIALTAIRGIGRRISTLICKKADIDTDKRAGELTADEISKIVAIVA 77
+RV N V + + L + GIGRR S + I T+K A +LT E+
Sbjct 32 IRVGNAEVPNNKPLKTGLQEVYGIGRRKSHQVLCHLGI-TNKLARDLTGKELI------- 83
Query 78 SPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRHYWGLKVR 137
D+++ +H + L + +RL ++ +RG RH GL R
Sbjct 84 ----------------DLREEVGQHQHGDELRRRVGSEIQRLVEVDCYRGSRHRHGLPCR 127
Query 138 GQHTKTTGR--HGRTVGVAKKK 157
GQ T T R G+ V +A KK
Sbjct 128 GQRTSTNARTKKGKAVAIAGKK 149
> YNL081c
Length=143
Score = 38.1 bits (87), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 32/134 (23%), Positives = 58/134 (43%), Gaps = 24/134 (17%)
Query 17 ILRVLNTNVDGREKVVIALTA-IRGIGRRISTLICKKADIDTDKRAGELTADEISKIVAI 75
++ +L G+E + IAL + GIG+ + IC K R +L+ +I I +
Sbjct 2 VVHILGKGFKGKEVIKIALASKFYGIGKTTAEKICSKLGFYPWMRMHQLSEPQIMSIASE 61
Query 76 VASPTQFSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLKKMRLHRGLRHYWGLK 135
+++ T +G +R + +N+ +K+ + G+RH L
Sbjct 62 LSTMTI---------------EGDARAIVKDNI--------ALKRKIGSYSGMRHTLHLP 98
Query 136 VRGQHTKTTGRHGR 149
VRGQHT+ + R
Sbjct 99 VRGQHTRNNAKTAR 112
> At5g14320
Length=197
Score = 37.0 bits (84), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 15/31 (48%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 116 FERLKKMRLHRGLRHYWGLKVRGQHTKTTGR 146
+RLK+++ +RG+RH GL RGQ TK R
Sbjct 150 IKRLKEIQCYRGVRHIQGLPCRGQRTKNNCR 180
> At3g01780
Length=1192
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 20/69 (28%), Positives = 33/69 (47%), Gaps = 1/69 (1%)
Query 17 ILRVLNTNVDGREKVVIALTAIRGIGRRISTLICKKADIDTDKRAGELTADEISKIVAIV 76
+L+ L + GR+ VIA +A+ I ++ +CKK D R+ LT D + + V
Sbjct 24 LLQALQQSAAGRDISVIAKSAVEEIVASPASAVCKKLAFDL-IRSTRLTPDLWDTVCSGV 82
Query 77 ASPTQFSIP 85
+ F P
Sbjct 83 KTDLHFPDP 91
> At4g18810
Length=621
Score = 28.1 bits (61), Expect = 7.6, Method: Composition-based stats.
Identities = 25/99 (25%), Positives = 46/99 (46%), Gaps = 7/99 (7%)
Query 27 GREKVVIALTAIRGIGRRISTLICK-----KADIDTDKRAGELTADEISKIVAIVASPTQ 81
G +++ A G+GRRI ++ K KA + +++A ++ EI IVA +
Sbjct 120 GTSGIILVAGATGGVGRRIVDILRKRGLPVKALVRNEEKARKMLGPEIDLIVADITKENT 179
Query 82 FSIPSWFLNRQKDVKDGKSRHVSANNLDSVWREDFERLK 120
+P F +K V + S V D+ R+ + ++K
Sbjct 180 L-VPEKFKGVRK-VINAVSVIVGPKEGDTPERQKYNQIK 216
> Hs4507857
Length=1102
Score = 28.1 bits (61), Expect = 7.9, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 26/50 (52%), Gaps = 2/50 (4%)
Query 13 DFQHILRVLNTNVDGREKVVIALTAIRGI--GRRISTLICKKADIDTDKR 60
D Q + RVL NV+ + K I + G+ +S + CK+ D +D+R
Sbjct 295 DVQELCRVLLDNVENKMKGTCVEGTIPKLFRGKMVSYIQCKEVDYRSDRR 344
Lambda K H
0.320 0.133 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2100092188
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40