bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3047_orf2
Length=219
Score E
Sequences producing significant alignments: (Bits) Value
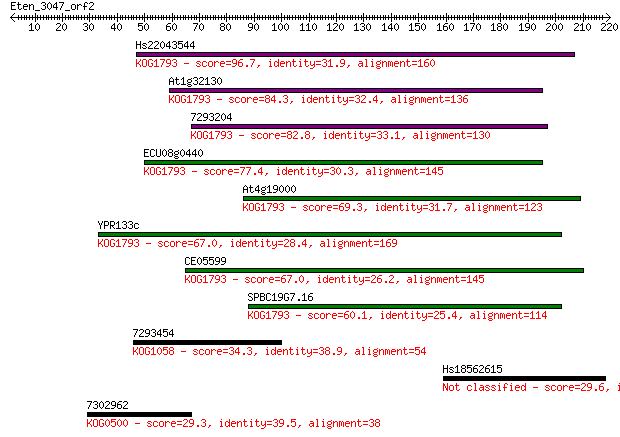
Hs22043544 96.7 3e-20
At1g32130 84.3 2e-16
7293204 82.8 5e-16
ECU08g0440 77.4 2e-14
At4g19000 69.3 5e-12
YPR133c 67.0 3e-11
CE05599 67.0 3e-11
SPBC19G7.16 60.1 4e-09
7293454 34.3 0.23
Hs18562615 29.6 4.7
7302962 29.3 6.8
> Hs22043544
Length=819
Score = 96.7 bits (239), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 51/161 (31%), Positives = 89/161 (55%), Gaps = 1/161 (0%)
Query 47 RQRRKRRPKPLDSDLQLFCNQMLQKMSEASQQDERSLEAGEPATAKLQMLDTVCTELVKP 106
++RR R SD + M+ KM+EA+++D + +PA KL +L V L K
Sbjct 542 KRRRNRDGGTFISDADDVVSAMIVKMNEAAEEDRQLNNQKKPALKKLTLLPAVVMHLKKQ 601
Query 107 KWRHWFVSEGCCQVIAGWLAPFKDDSLPNFSLRRRLLEVLGQMPITAQD-LANNDLGKVL 165
+ F+ G I WL+P D SLP +R LL++L ++P +Q+ L ++ +G+ +
Sbjct 602 DLKETFIDSGVMSAIKEWLSPLPDRSLPALKIREELLKILQELPSVSQETLKHSGIGRAV 661
Query 166 VLLWQHDDETESNRQLIRGLIQKWMRPVLGLGSSHRQLISE 206
+ L++H E+ SN+ + LI +W RP+ GL S+++ + E
Sbjct 662 MYLYKHPKESRSNKDMAGKLINEWSRPIFGLTSNYKGMTRE 702
> At1g32130
Length=404
Score = 84.3 bits (207), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 44/142 (30%), Positives = 75/142 (52%), Gaps = 6/142 (4%)
Query 59 SDLQLFCNQMLQKMSEASQQDERSLEAGEPATAKLQMLDTVCTELVKPKWRHWFVSEGCC 118
+++ L ++ ++ +++D G+PA KL+ L + L K + + F+ G
Sbjct 129 AEIALLVENVMAELEVTAEEDAELNRQGKPAINKLKKLSLLTDVLGKKQLQTEFLDHGVL 188
Query 119 QVIAGWLAPFKDDSLPNFSLRRRLLEVLGQMPIT------AQDLANNDLGKVLVLLWQHD 172
++ WL P D SLPN ++R +L VL PI + L + LGKV++ L + D
Sbjct 189 TLLKNWLEPLPDGSLPNINIRAAILRVLTDFPIDLDQYDRREQLKKSGLGKVIMFLSKSD 248
Query 173 DETESNRQLIRGLIQKWMRPVL 194
+ET SNR+L + L+ KW RP+
Sbjct 249 EETNSNRRLAKDLVDKWSRPIF 270
> 7293204
Length=797
Score = 82.8 bits (203), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 43/131 (32%), Positives = 70/131 (53%), Gaps = 1/131 (0%)
Query 67 QMLQKMSEASQQDERSLEAGEPATAKLQMLDTVCTELVKPKWRHWFVSEGCCQVIAGWLA 126
Q++ M AS D + G+PAT K+ ML V ++L+K + F+ V+ WLA
Sbjct 540 QLIVSMKNASDDDRQLNMIGQPATKKISMLKQVMSQLIKKHLQLAFLEHNILNVLTDWLA 599
Query 127 PFKDDSLPNFSLRRRLLEVLGQMPITAQD-LANNDLGKVLVLLWQHDDETESNRQLIRGL 185
P + SLP +R +L++L P + L + +GK ++ L++H ET+SNR L
Sbjct 600 PLPNKSLPCLQIRESILKLLSDFPTIEKGLLKQSGIGKAVMYLYKHPQETKSNRDRAGRL 659
Query 186 IQKWMRPVLGL 196
I +W RP+ +
Sbjct 660 ISEWARPIFNV 670
> ECU08g0440
Length=198
Score = 77.4 bits (189), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 44/146 (30%), Positives = 73/146 (50%), Gaps = 1/146 (0%)
Query 50 RKRRPKPLDSDLQLFCNQMLQK-MSEASQQDERSLEAGEPATAKLQMLDTVCTELVKPKW 108
R R LD L ++ L+K M + ++D + G PAT K++ ++ + L+
Sbjct 47 RAERKGDLDPGTVLEISRSLKKRMQDILKKDNANNLEGRPATGKIENVEEISDILMSKAL 106
Query 109 RHWFVSEGCCQVIAGWLAPFKDDSLPNFSLRRRLLEVLGQMPITAQDLANNDLGKVLVLL 168
+ + EG I GWL P D S+PN +R+RLL+VL M I + L + +GK++
Sbjct 107 QESLLDEGILDEIKGWLEPLPDKSMPNIKIRKRLLDVLKTMKIHKEHLVTSGVGKIVYFY 166
Query 169 WQHDDETESNRQLIRGLIQKWMRPVL 194
+ E++ R + L+QKW V
Sbjct 167 SINPKESKEVRASAKALVQKWTNEVF 192
> At4g19000
Length=406
Score = 69.3 bits (168), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 39/129 (30%), Positives = 63/129 (48%), Gaps = 6/129 (4%)
Query 86 GEPATAKLQMLDTVCTELVKPKWRHWFVSEGCCQVIAGWLAPFKDDSLPNFSLRRRLLEV 145
G+PA KL L + L K + F+ G ++ WL P D SLPN ++R +L +
Sbjct 168 GKPAINKLMKLPLLNETLSKKPLQGEFLDHGVLNLLKNWLEPLPDGSLPNINIRSAVLMI 227
Query 146 LGQMPIT------AQDLANNDLGKVLVLLWQHDDETESNRQLIRGLIQKWMRPVLGLGSS 199
L I + L + LGKV++ L + D+ET NR+L +I KW R + +
Sbjct 228 LNDFRIDLDQDSRREQLIKSGLGKVIMFLSKSDEETTPNRRLANDIINKWGRIIYNKSTR 287
Query 200 HRQLISERE 208
+ + ++ E
Sbjct 288 YDNMFTQEE 296
> YPR133c
Length=410
Score = 67.0 bits (162), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 48/184 (26%), Positives = 90/184 (48%), Gaps = 15/184 (8%)
Query 33 SADPTS---FESALNRLRQRRK-RRPKPLDSDLQLFCNQML----QKMSEASQQD----E 80
SA P+S E L+R+ ++ K RR + + DL+ + ++ + +M+ A+Q D
Sbjct 118 SATPSSRQELEEKLDRILKKPKVRRTRRDEDDLEQYLDEKILRLKDEMNIAAQLDIDTLN 177
Query 81 RSLEAGEP---ATAKLQMLDTVCTELVKPKWRHWFVSEGCCQVIAGWLAPFKDDSLPNFS 137
+ +E G+ A K+++L V + L K + Q + WL P D SLP+F
Sbjct 178 KRIETGDTSLIAMQKVKLLPKVVSVLSKANLADTILDNNLLQSVRIWLEPLPDGSLPSFE 237
Query 138 LRRRLLEVLGQMPITAQDLANNDLGKVLVLLWQHDDETESNRQLIRGLIQKWMRPVLGLG 197
+++ L L +P+ + L + LG+V++ + +L LI +W RP++G
Sbjct 238 IQKSLFAALNDLPVKTEHLKESGLGRVVIFYTKSKRVEAQLARLAEKLIAEWTRPIIGAS 297
Query 198 SSHR 201
++R
Sbjct 298 DNYR 301
> CE05599
Length=511
Score = 67.0 bits (162), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 38/148 (25%), Positives = 77/148 (52%), Gaps = 4/148 (2%)
Query 65 CNQMLQKMSEASQQDERSLEAGEPATAKLQMLDTVCTELVKPKWRHWFVSEGCCQVIAGW 124
++++++M A++ D + +PA K++ML V +++ + G ++ W
Sbjct 240 VSRLVERMKHAAKSDRNANIERKPAFQKIKMLPEVKAIMLRAGIVEVLIENGFMSALSEW 299
Query 125 LAPFKDDSLPNFSLRRRLLEVLGQM---PITAQDLANNDLGKVLVLLWQHDDETESNRQL 181
LAP D LP +R +L++L + L + LGK +++L++H +ET+ N+ +
Sbjct 300 LAPLPDKCLPALDIRITVLKLLHNPRFWKLDRSTLKQSGLGKAVMMLYKHPNETKENKGI 359
Query 182 IRGLIQKWMRPVLGLGSSHRQLISERER 209
LI +W RP+ L + + +S +ER
Sbjct 360 ANKLIGEWARPIYHLDTDY-STVSRQER 386
> SPBC19G7.16
Length=428
Score = 60.1 bits (144), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 29/114 (25%), Positives = 57/114 (50%), Gaps = 0/114 (0%)
Query 88 PATAKLQMLDTVCTELVKPKWRHWFVSEGCCQVIAGWLAPFKDDSLPNFSLRRRLLEVLG 147
PAT KL+ML V L K + + WL P D SLP +++R L+++L
Sbjct 212 PATEKLKMLPLVDAVLRKTHLYDTILDNNVLDSVRMWLEPLPDRSLPALNIQRSLMDILT 271
Query 148 QMPITAQDLANNDLGKVLVLLWQHDDETESNRQLIRGLIQKWMRPVLGLGSSHR 201
++PI + L + +G++++ +++ L+ +W RP++ +++R
Sbjct 272 KLPIQTEHLRESKIGRIVLFYTISKKPEPFIKRIADNLVSEWSRPIIKRSANYR 325
> 7293454
Length=963
Score = 34.3 bits (77), Expect = 0.23, Method: Composition-based stats.
Identities = 21/55 (38%), Positives = 32/55 (58%), Gaps = 2/55 (3%)
Query 46 LRQRRKRRPKPLDSDLQLFCNQMLQKMSEASQ-QDERSLEAGEPATAKLQMLDTV 99
LR +R P+ + S L+C + L KM +A +D+R L+ + ATAK+Q D V
Sbjct 629 LRTLSERTPEAI-SVFTLYCREALGKMLDAQHDEDQRMLKEKQKATAKVQPDDPV 682
> Hs18562615
Length=697
Score = 29.6 bits (65), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 33/60 (55%), Gaps = 8/60 (13%)
Query 159 NDLGKVLVLLWQHDDETESN-RQLIRGLIQKWMRPVLGLGSSHRQLISERERAFEANDSD 217
+DL K LV D+TE ++L + L L S RQL++ER+RA+EA+D +
Sbjct 219 DDLAKKLVSAELKLDDTERRIKELSKNL-------ELSTNSFQRQLLAERKRAYEAHDEN 271
> 7302962
Length=665
Score = 29.3 bits (64), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 23/40 (57%), Gaps = 2/40 (5%)
Query 29 DPAPSADPTSFESALNRLRQR--RKRRPKPLDSDLQLFCN 66
+P+ + P++ L LRQR ++ RPKP D L+ F N
Sbjct 26 EPSKRSKPSALRRTLQALRQRLTKRNRPKPPDWFLEKFSN 65
Lambda K H
0.321 0.134 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4112032690
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40