bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3093_orf1
Length=264
Score E
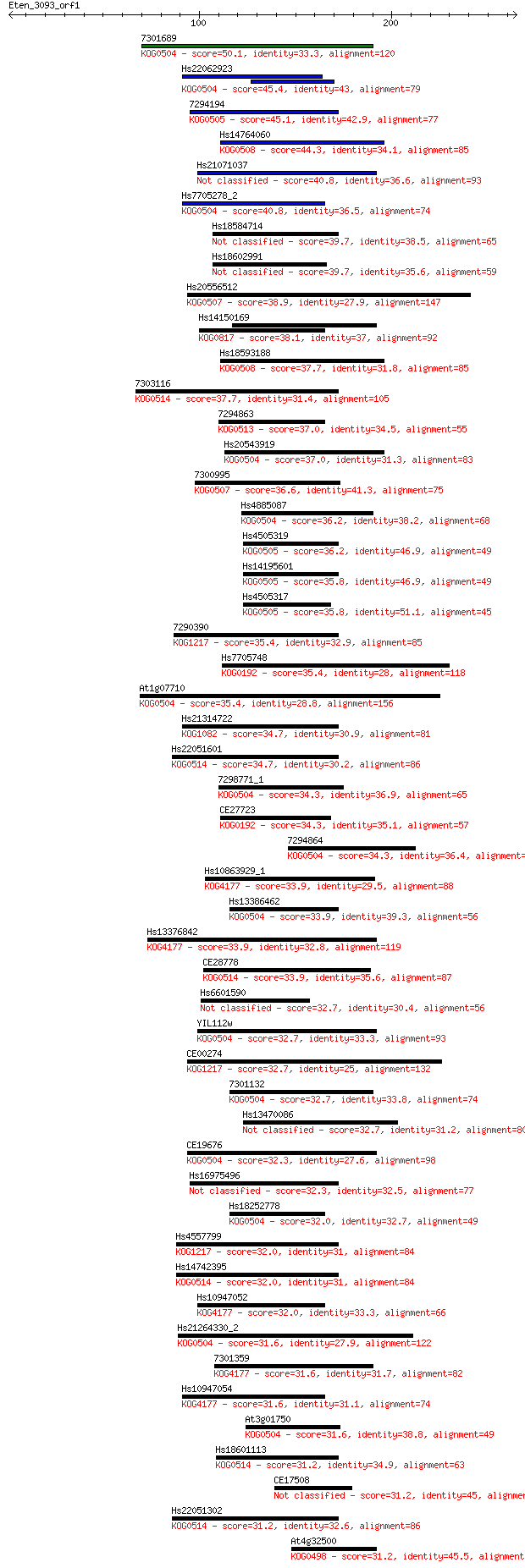
Sequences producing significant alignments: (Bits) Value
7301689 50.1 4e-06
Hs22062923 45.4 1e-04
7294194 45.1 1e-04
Hs14764060 44.3 3e-04
Hs21071037 40.8 0.003
Hs7705278_2 40.8 0.003
Hs18584714 39.7 0.006
Hs18602991 39.7 0.007
Hs20556512 38.9 0.011
Hs14150169 38.1 0.019
Hs18593188 37.7 0.022
7303116 37.7 0.027
7294863 37.0 0.039
Hs20543919 37.0 0.041
7300995 36.6 0.062
Hs4885087 36.2 0.071
Hs4505319 36.2 0.081
Hs14195601 35.8 0.085
Hs4505317 35.8 0.091
7290390 35.4 0.11
Hs7705748 35.4 0.12
At1g07710 35.4 0.13
Hs21314722 34.7 0.21
Hs22051601 34.7 0.23
7298771_1 34.3 0.25
CE27723 34.3 0.26
7294864 34.3 0.30
Hs10863929_1 33.9 0.36
Hs13386462 33.9 0.36
Hs13376842 33.9 0.36
CE28778 33.9 0.40
Hs6601590 32.7 0.70
YIL112w 32.7 0.80
CE00274 32.7 0.82
7301132 32.7 0.85
Hs13470086 32.7 0.86
CE19676 32.3 1.0
Hs16975496 32.3 1.2
Hs18252778 32.0 1.3
Hs4557799 32.0 1.3
Hs14742395 32.0 1.4
Hs10947052 32.0 1.4
Hs21264330_2 31.6 1.7
7301359 31.6 1.7
Hs10947054 31.6 1.8
At3g01750 31.6 1.8
Hs18601113 31.2 2.1
CE17508 31.2 2.1
Hs22051302 31.2 2.3
At4g32500 31.2 2.3
> 7301689
Length=2119
Score = 50.1 bits (118), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 40/122 (32%), Positives = 64/122 (52%), Gaps = 10/122 (8%)
Query 70 GSLGTVKAMKRYWNQPVSDMNDDSNASKYTLLQMALMLQKKKVVEYLLPQQDLDLNTKAT 129
G + TV+ + ++ D+N S+ TL +AL K ++V+YLL ++D+N +
Sbjct 1629 GHMDTVEFLLKF----CCDVNSKDADSRTTLYILALE-NKLEIVKYLLDMTNVDVNIPDS 1683
Query 130 DGTTALMTACDQQVPEDWVQAMLERGANKSINDEDNEGRTALDRC--EKGSPLYLLLLKY 187
+G TAL A Q D V+ ++E GA+ +N D E RT L C + + +LL Y
Sbjct 1684 EGRTALHVAA-WQGHADMVKTLIEAGAD--VNSMDLEARTPLHSCAWQGNHDVMNILLYY 1740
Query 188 GA 189
GA
Sbjct 1741 GA 1742
> Hs22062923
Length=794
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/73 (39%), Positives = 40/73 (54%), Gaps = 2/73 (2%)
Query 91 DDSNASKYTLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQA 150
+D+ + TLL MA+ Q K +LL Q D+N + DG TAL A Q+P V A
Sbjct 241 NDTMSDGQTLLHMAIQRQDSKSALFLLEHQA-DINVRTQDGETALQLAIRNQLPL-VVDA 298
Query 151 MLERGANKSINDE 163
+ RGA+ S+ DE
Sbjct 299 ICTRGADMSVPDE 311
Score = 29.6 bits (65), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 20/43 (46%), Positives = 26/43 (60%), Gaps = 3/43 (6%)
Query 127 KATDGTTALMTACDQQVPEDWVQAMLERGANKSINDEDNEGRT 169
+A DG T L A + E+ VQ +LE GAN +N +D EGRT
Sbjct 391 EARDGQTPLHLAASWGL-EETVQCLLEFGAN--VNAQDAEGRT 430
> 7294194
Length=1023
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/78 (42%), Positives = 43/78 (55%), Gaps = 7/78 (8%)
Query 95 ASKYTLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTAC-DQQVPEDWVQAMLE 153
+S L L K +VV+ L Q D+NT DG TAL AC D + D V+ ++E
Sbjct 45 SSGCVFLAACLSGDKDEVVQLL--DQGADINTANVDGLTALHQACIDDNL--DMVEFLVE 100
Query 154 RGANKSINDEDNEGRTAL 171
RGA+ IN +DNEG T L
Sbjct 101 RGAD--INRQDNEGWTPL 116
> Hs14764060
Length=669
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/85 (34%), Positives = 42/85 (49%), Gaps = 3/85 (3%)
Query 111 KVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLERGANKSINDEDNEGRTA 170
+VV YL+ + DL G T LM +C + E + +LE+GA +N +G TA
Sbjct 130 EVVRYLVGEHQADLEVANRHGHTCLMISCYKGHRE-IARYLLEQGAQ--VNRRSAKGNTA 186
Query 171 LDRCEKGSPLYLLLLKYGAKPRPEK 195
L C + L +L L G K R E+
Sbjct 187 LHDCAESGSLEILQLLLGCKARMER 211
> Hs21071037
Length=1721
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 34/97 (35%), Positives = 47/97 (48%), Gaps = 9/97 (9%)
Query 99 TLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDW---VQAMLERG 155
TLLQ A L ++VV Y L + D+N + G AL AC + W V+ +LE G
Sbjct 1431 TLLQRAARLGYEEVVLYCLENKICDVNHRDNAGYCALHEACAR----GWLNIVRHLLEYG 1486
Query 156 ANKSINDEDNEGRTALDRCEKGS-PLYLLLLKYGAKP 191
A+ + + +D R D E + LLL YGA P
Sbjct 1487 ADVNCSAQDGT-RPLHDAVENDHLEIVRLLLSYGADP 1522
> Hs7705278_2
Length=860
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 40/74 (54%), Gaps = 2/74 (2%)
Query 91 DDSNASKYTLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQA 150
+D+ + TLL MA+ Q K +LL + D+N + G TAL A Q+P V A
Sbjct 308 NDTMSDGQTLLHMAIQRQDSKSALFLLEHRA-DINVRPQAGETALQLAIRNQLPL-VVDA 365
Query 151 MLERGANKSINDED 164
+ RGA+ S+ DE+
Sbjct 366 ICTRGADMSVPDEE 379
> Hs18584714
Length=535
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 38/65 (58%), Gaps = 3/65 (4%)
Query 107 LQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLERGANKSINDEDNE 166
+ K K+V+YLL + D N + G TAL+ AC ++ + V +LE GA+ S+ ED
Sbjct 62 ISKSKMVKYLLDNR-ADPNIQDKSGKTALIHACIRRAGGEVVSLLLENGADPSL--EDRT 118
Query 167 GRTAL 171
G +AL
Sbjct 119 GASAL 123
> Hs18602991
Length=980
Score = 39.7 bits (91), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 35/59 (59%), Gaps = 1/59 (1%)
Query 107 LQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLERGANKSINDEDN 165
+ K K+V+YLL + + D N + G TALM AC ++ + V +L+ GA+ S+ D +
Sbjct 479 VSKAKMVKYLL-ENNADPNIQDKSGKTALMHACLEKAGPEVVSLLLKSGADLSLQDHSS 536
> Hs20556512
Length=765
Score = 38.9 bits (89), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 41/154 (26%), Positives = 67/154 (43%), Gaps = 12/154 (7%)
Query 94 NASKYTLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLE 153
N K+T L +A K VV+ LL +D N + G+ A + D VQ +L
Sbjct 306 NTKKHTPLHLAARNGHKAVVQVLL-DAGMDSNYQTEMGSALHEAALFGKT--DVVQILLA 362
Query 154 RGANKSINDEDNEGRTALDRC-----EKGSPLYLLLLKY--GAKPRPEKLPTPPPEPEVT 206
G + +N +DN G TALD +K + L+ + G + E TPPP+P +
Sbjct 363 AGTD--VNIKDNHGLTALDTVRELPSQKSQQIAALIEDHMTGKRSTKEVDKTPPPQPPLI 420
Query 207 VAASNPPASTDSEATKQTPEIVMADDVPGLEQHP 240
+ + + + K E+++ D E+ P
Sbjct 421 SSMDSISQKSQGDVEKAVTELIIDFDANAEEEGP 454
> Hs14150169
Length=282
Score = 38.1 bits (87), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 28/77 (36%), Positives = 42/77 (54%), Gaps = 5/77 (6%)
Query 117 LPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLERGANKSINDEDNEGRTALDRCEK 176
+ +++D+N K +G L ACD+ E V +L+ A+ IN +DNEG+TAL
Sbjct 178 IKSKNVDVNVKDEEGRALLHWACDRGHKE-LVTVLLQHRAD--INCQDNEGQTALHYASA 234
Query 177 GSPLYL--LLLKYGAKP 191
L + LLL+ GA P
Sbjct 235 CEFLDIVELLLQSGADP 251
Score = 30.8 bits (68), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 37/67 (55%), Gaps = 6/67 (8%)
Query 100 LLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALM--TACDQQVPEDWVQAMLERGAN 157
LL A K++V LL Q D+N + +G TAL +AC+ D V+ +L+ GA+
Sbjct 195 LLHWACDRGHKELVTVLL-QHRADINCQDNEGQTALHYASACEFL---DIVELLLQSGAD 250
Query 158 KSINDED 164
++ D+D
Sbjct 251 PTLRDQD 257
> Hs18593188
Length=1172
Score = 37.7 bits (86), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 27/85 (31%), Positives = 41/85 (48%), Gaps = 3/85 (3%)
Query 111 KVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLERGANKSINDEDNEGRTA 170
+VV YL+ + +L G LM +C + E + +LE+GA +N +G TA
Sbjct 634 EVVRYLVGEHQANLEVANRHGHMCLMISCYKGHRE-IARYLLEQGAQ--VNWRSAKGNTA 690
Query 171 LDRCEKGSPLYLLLLKYGAKPRPEK 195
L C + S L +L L G K E+
Sbjct 691 LHNCAETSSLEILQLLLGCKASMER 715
> 7303116
Length=1224
Score = 37.7 bits (86), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 33/109 (30%), Positives = 54/109 (49%), Gaps = 11/109 (10%)
Query 67 IANGSLGTVKAMKRYWNQPVSDMNDDSNASKYTLLQMALMLQK----KKVVEYLLPQQDL 122
+++G+ V + + V D+N +NA +++ ++L K + VVE L D+
Sbjct 1040 VSHGNFDVVSIL---LDSKVCDVNQMNNAGYTSVMLVSLAKLKQPAHRTVVERLFKMADV 1096
Query 123 DLNTKATDGTTALMTACDQQVPEDWVQAMLERGANKSINDEDNEGRTAL 171
++ K TALM A D V +L+ GA+ +I DED G TAL
Sbjct 1097 NIRAKK-HCQTALMLAVSHG-NGDMVNMLLDAGADINIQDED--GSTAL 1141
> 7294863
Length=877
Score = 37.0 bits (84), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 34/55 (61%), Gaps = 1/55 (1%)
Query 110 KKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLERGANKSINDED 164
K+++ ++ + ++LN +DG T L AC PE+ V+A+L GAN ++N +D
Sbjct 198 KEIINLIIDKSTVNLNHLNSDGYTPLHVACLADKPEN-VKALLLAGANVNLNAKD 251
> Hs20543919
Length=339
Score = 37.0 bits (84), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 41/83 (49%), Gaps = 1/83 (1%)
Query 113 VEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLERGANKSINDEDNEGRTALD 172
V LL ++N K +G T LM A E+ VQ +L++GA+ S+ +E +G +
Sbjct 254 VASLLIDAGANVNVKDRNGKTPLMVAVLNN-HEELVQLLLDKGADASVKNEFGKGVLEMA 312
Query 173 RCEKGSPLYLLLLKYGAKPRPEK 195
R + LL + K RP+K
Sbjct 313 RVFDRQSVVSLLEERKKKQRPKK 335
> 7300995
Length=1315
Score = 36.6 bits (83), Expect = 0.062, Method: Compositional matrix adjust.
Identities = 31/77 (40%), Positives = 41/77 (53%), Gaps = 9/77 (11%)
Query 98 YTLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTA--CDQQVPEDWVQAMLERG 155
+T L +A K VVE LL + +N GT AL A C + E+ V+ +L+ G
Sbjct 206 HTCLHLASRNGHKSVVEVLL-SAGVSVNLLTPSGT-ALHEAALCGK---ENVVRTLLKAG 260
Query 156 ANKSINDEDNEGRTALD 172
N +N DNEGRTALD
Sbjct 261 IN--LNATDNEGRTALD 275
> Hs4885087
Length=446
Score = 36.2 bits (82), Expect = 0.071, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 39/70 (55%), Gaps = 4/70 (5%)
Query 122 LDLNTKATDGTTALMTACDQQVPEDWVQAMLERGANKSINDEDNEGRTALDRCEKGSPLY 181
LDL + DG TAL A + + ++ VQ +LERGA+ D GR+ L + + L
Sbjct 225 LDLEARNYDGLTALHVAVNTEC-QETVQLLLERGADIDAVDI-KSGRSPLIHAVENNSLS 282
Query 182 L--LLLKYGA 189
+ LLL++GA
Sbjct 283 MVQLLLQHGA 292
> Hs4505319
Length=982
Score = 36.2 bits (82), Expect = 0.081, Method: Compositional matrix adjust.
Identities = 23/50 (46%), Positives = 31/50 (62%), Gaps = 5/50 (10%)
Query 123 DLNTKATDGTTALMTAC-DQQVPEDWVQAMLERGANKSINDEDNEGRTAL 171
D+NT DG TAL AC D+ + D V+ ++E AN +N +DNEG T L
Sbjct 83 DINTVNVDGLTALHQACIDENL--DMVKFLVENRAN--VNQQDNEGWTPL 128
> Hs14195601
Length=998
Score = 35.8 bits (81), Expect = 0.085, Method: Compositional matrix adjust.
Identities = 23/50 (46%), Positives = 31/50 (62%), Gaps = 5/50 (10%)
Query 123 DLNTKATDGTTALMTAC-DQQVPEDWVQAMLERGANKSINDEDNEGRTAL 171
D+NT DG TAL AC D+ + D V+ ++E AN +N +DNEG T L
Sbjct 83 DINTVNVDGLTALHQACIDENL--DMVKFLVENRAN--VNQQDNEGWTPL 128
> Hs4505317
Length=1030
Score = 35.8 bits (81), Expect = 0.091, Method: Compositional matrix adjust.
Identities = 23/46 (50%), Positives = 27/46 (58%), Gaps = 5/46 (10%)
Query 123 DLNTKATDGTTALMTAC-DQQVPEDWVQAMLERGANKSINDEDNEG 167
D+N DG TAL AC D V D V+ ++E GAN IN DNEG
Sbjct 65 DINYANVDGLTALHQACIDDNV--DMVKFLVENGAN--INQPDNEG 106
> 7290390
Length=2634
Score = 35.4 bits (80), Expect = 0.11, Method: Composition-based stats.
Identities = 28/85 (32%), Positives = 42/85 (49%), Gaps = 4/85 (4%)
Query 87 SDMNDDSNASKYTLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPED 146
+D N N + T L A+ V + LL + +LN + DGTT L+ A + E
Sbjct 1975 ADANCQDNTGR-TPLHAAVAADAMGVFQILLRNRATNLNARMHDGTTPLILAARLAI-EG 2032
Query 147 WVQAMLERGANKSINDEDNEGRTAL 171
V+ ++ A+ IN DN G+TAL
Sbjct 2033 MVEDLIT--ADADINAADNSGKTAL 2055
> Hs7705748
Length=835
Score = 35.4 bits (80), Expect = 0.12, Method: Composition-based stats.
Identities = 33/129 (25%), Positives = 60/129 (46%), Gaps = 13/129 (10%)
Query 112 VVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLERGANKSI--------NDE 163
+V++LL Q +++N + DG T L +AC VQ +L+ GA+ ++ + E
Sbjct 321 LVKFLLDQNVININHQGRDGHTGLHSACYHGHIR-LVQFLLDNGADMNLVACDPSRSSGE 379
Query 164 DNEGRTALDRCEKGSPLYLLLLKYGAKPRPEKLPT---PPPEPEVTVAASNPPASTDSEA 220
+E + EKG + LLK+ +P+ E LP P + + + P
Sbjct 380 KDEQTCLMWAYEKGHDAIVTLLKHYKRPQDE-LPCNEYSQPGGDGSYVSVPSPLGKIKSM 438
Query 221 TKQTPEIVM 229
TK+ +I++
Sbjct 439 TKEKADILL 447
> At1g07710
Length=543
Score = 35.4 bits (80), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 45/163 (27%), Positives = 69/163 (42%), Gaps = 15/163 (9%)
Query 69 NGSLGTVKAMKRYWNQPVSDMNDDSNASKYTLLQMALMLQKKKVVEYLLPQQDLDLNTKA 128
NG + +KA+ ++P + D T L MA+ +VVE L+ +N
Sbjct 179 NGHVKVIKAL--LASEPAIAIRMDKKGQ--TALHMAVKGTNVEVVEELIKADRSSINIAD 234
Query 129 TDGTTALMTACDQQVPEDWVQAMLERGA--NKSINDEDNEGRTALDRCEK-GSPLYLLLL 185
T G TAL A ++ V+ +L K++N G TALD EK G+P L+L
Sbjct 235 TKGNTALHIAA-RKGRSQIVKLLLANNMTDTKAVN---RSGETALDTAEKIGNPEVALIL 290
Query 186 KYGAKPRPEKL----PTPPPEPEVTVAASNPPASTDSEATKQT 224
+ P + + P P E + TV+ E T+ T
Sbjct 291 QKHGVPSAKTIKPSGPNPARELKQTVSDIKHEVHNQLEHTRLT 333
> Hs21314722
Length=1267
Score = 34.7 bits (78), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 41/81 (50%), Gaps = 3/81 (3%)
Query 91 DDSNASKYTLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQA 150
D +A T L +A +VV+YLL +D+N + G T ++ A + + D V+
Sbjct 802 DPKDAEGSTCLHLAAKKGHYEVVQYLLSNGQMDVNCQDDGGWTPMIWATEYK-HVDLVKL 860
Query 151 MLERGANKSINDEDNEGRTAL 171
+L +G++ IN DNE L
Sbjct 861 LLSKGSD--INIRDNEENICL 879
> Hs22051601
Length=454
Score = 34.7 bits (78), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 46/86 (53%), Gaps = 3/86 (3%)
Query 86 VSDMNDDSNASKYTLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPE 145
+ ++N ++ + T L +A+ + VV+ LL + D+N + DG+TALM AC+ E
Sbjct 341 LGNINAKASQAGQTALMLAVSHGRVDVVKALLACE-ADVNVQDDDGSTALMCACEHGHKE 399
Query 146 DWVQAMLERGANKSINDEDNEGRTAL 171
+ +L + I+ D +G TAL
Sbjct 400 --IAGLLLAVPSCDISLTDRDGSTAL 423
> 7298771_1
Length=481
Score = 34.3 bits (77), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 35/67 (52%), Gaps = 3/67 (4%)
Query 110 KKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLERGANKSINDEDNE--G 167
++++E L DLD+N TTAL A E V+ +LERGA + DEDN+
Sbjct 60 QRILERFLECPDLDVNALCDGQTTALHIASIYGRTE-LVRMLLERGACADLADEDNKLPV 118
Query 168 RTALDRC 174
A++ C
Sbjct 119 HYAIEEC 125
> CE27723
Length=850
Score = 34.3 bits (77), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 31/57 (54%), Gaps = 1/57 (1%)
Query 111 KVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLERGANKSINDEDNEG 167
++V +L+ +D N + DG TAL +AC VQ +LE GA++S+ EG
Sbjct 336 ELVSFLMKYPGVDPNYQGQDGHTALHSACYHGHLR-IVQYLLENGADQSLASRAFEG 391
> 7294864
Length=358
Score = 34.3 bits (77), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 31/67 (46%), Gaps = 1/67 (1%)
Query 146 DWVQAMLERGANKSINDEDNEGRTALDRCEKGSPLYLLLLKYGAKPR-PEKLPTPPPEPE 204
+ + +LE GAN + DE N L C P+ LLKYGA P + L P
Sbjct 177 ELLNRILEGGANPNAADEYNRSPLHLAACRGYIPIVQQLLKYGANPNVVDSLGNTPLHLA 236
Query 205 VTVAASN 211
V A+SN
Sbjct 237 VISASSN 243
> Hs10863929_1
Length=361
Score = 33.9 bits (76), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 26/90 (28%), Positives = 47/90 (52%), Gaps = 8/90 (8%)
Query 103 MALMLQKKKV--VEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLERGANKSI 160
+ L ++KK + V+ LL Q+ +++N +DG TAL+ A + ++ + + + +RGA+
Sbjct 243 LILAVEKKHLGLVQRLLEQEHIEINDTDSDGKTALLLAVELKLKK-IAELLCKRGASTDC 301
Query 161 NDEDNEGRTALDRCEKGSPLYLLLLKYGAK 190
D R D L +LL +GAK
Sbjct 302 GDLVMTARRNYDH-----SLVKVLLSHGAK 326
> Hs13386462
Length=435
Score = 33.9 bits (76), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 29/56 (51%), Gaps = 3/56 (5%)
Query 116 LLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLERGANKSINDEDNEGRTAL 171
LL D D+N+++ G TAL AC D V+ +L GAN I D + G T L
Sbjct 98 LLLLHDADVNSQSATGNTALTYACAGGFV-DIVKVLLNEGAN--IEDHNENGHTPL 150
> Hs13376842
Length=1166
Score = 33.9 bits (76), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 39/121 (32%), Positives = 55/121 (45%), Gaps = 7/121 (5%)
Query 73 GTVKAMKRYWNQPVSDMNDDSNASKYTLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGT 132
G V+ +KR P + D+ K T L A +K VVEYLL Q ++ + G
Sbjct 35 GDVERVKRLVT-PEKVNSRDTAGRKSTPLHFAAGFGRKDVVEYLL-QNGANVQARDDGGL 92
Query 133 TALMTACDQQVPEDWVQAMLERGANKSINDEDNEGRTALDRCE-KGS-PLYLLLLKYGAK 190
L AC E V +L GA+ N DN T L KG + ++LL++GA+
Sbjct 93 IPLHNACSFGHAE-VVNLLLRHGADP--NARDNWNYTPLHEAAIKGKIDVCIVLLQHGAE 149
Query 191 P 191
P
Sbjct 150 P 150
> CE28778
Length=364
Score = 33.9 bits (76), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 31/94 (32%), Positives = 41/94 (43%), Gaps = 8/94 (8%)
Query 102 QMALML---QKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLERGANK 158
Q ALML KK LL ++N + DG+TALM A + E + E N
Sbjct 261 QTALMLAVSHGKKTTTELLLACGANVNEQDQDGSTALMCAAEHGHKELVKMLLAENLVNA 320
Query 159 SINDEDNEGRTAL----DRCEKGSPLYLLLLKYG 188
S+ D DN ++ D E G +Y L YG
Sbjct 321 SLTDVDNSTALSIALENDHREIGVMIY-AYLNYG 353
> Hs6601590
Length=1119
Score = 32.7 bits (73), Expect = 0.70, Method: Composition-based stats.
Identities = 17/56 (30%), Positives = 32/56 (57%), Gaps = 1/56 (1%)
Query 101 LQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLERGA 156
L +A+ +V++ LL + +D+N + +G TA++ AC E +Q +L +GA
Sbjct 135 LHIAVQGMNNEVMKVLLEHRTIDVNLEGENGNTAVIIACTTNNSEA-LQILLNKGA 189
> YIL112w
Length=1083
Score = 32.7 bits (73), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 31/95 (32%), Positives = 46/95 (48%), Gaps = 3/95 (3%)
Query 99 TLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLERGANK 158
T LQ+A K VV+ ++ + D+N + G TAL A Q E V+ ++E GA+
Sbjct 332 TRLQIACDKGKYDVVKKMIEEGGYDINDQDNAGNTALHEAALQGHIE-IVELLIENGADV 390
Query 159 SINDEDNEGRTALDRCEKGSPLYLL--LLKYGAKP 191
+I + G T L L ++ LLK GA P
Sbjct 391 NIKSIEMFGDTPLIDASANGHLDVVKYLLKNGADP 425
> CE00274
Length=1429
Score = 32.7 bits (73), Expect = 0.82, Method: Composition-based stats.
Identities = 33/149 (22%), Positives = 69/149 (46%), Gaps = 19/149 (12%)
Query 94 NASKY---TLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQA 150
++ KY T L A + +V+YL+ ++ + + + DG T +M A Q+ + V
Sbjct 1201 DSEKYKGRTALHYAAQVSNMPIVKYLVGEKGSNKDKQDEDGKTPIMLAA-QEGRIEVVMY 1259
Query 151 MLERGAN-KSINDEDNEGRT------------ALDRCEKGSPLYL-LLLKYGAKPRPEKL 196
++++GA+ ++++ D+ R DRC + L +++ +P+P +
Sbjct 1260 LIQQGASVEAVDATDHTARQLAQANNHHNIVDIFDRCRPEREYSMDLHIQHTHQPQPSRK 1319
Query 197 PTPPPEPEVTVAASNPPASTDSEATKQTP 225
T P+ + T + AS ++T TP
Sbjct 1320 VTRAPKKQ-TSRSKKESASNSRDSTHLTP 1347
> 7301132
Length=784
Score = 32.7 bits (73), Expect = 0.85, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 39/77 (50%), Gaps = 7/77 (9%)
Query 116 LLPQQDLDLNTKATDGTTALMTAC-DQQVPEDWVQAMLERGANKSINDEDNEGRTALDRC 174
LL + D+N G T LM AC QV D V+ +L+ GAN + +++ G T L
Sbjct 47 LLLNHNADVNAHCATGNTPLMFACAGGQV--DVVKVLLKHGAN--VEEQNENGHTPLMEA 102
Query 175 EKGSPLYL--LLLKYGA 189
+ + +LL++GA
Sbjct 103 ASAGHVEVAKVLLEHGA 119
> Hs13470086
Length=980
Score = 32.7 bits (73), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 45/82 (54%), Gaps = 5/82 (6%)
Query 123 DLNTKATDGTTALMTACDQQVPEDWVQAMLERGANKSINDEDNEGRTAL--DRCEKGSPL 180
D+N++ G TALM AC+ + V+A++++GA+ +N D+ G AL + + + +
Sbjct 177 DVNSRNKSGRTALMLACEIG-SSNAVEALIKKGAD--LNLVDSLGYNALHYSKLSENAGI 233
Query 181 YLLLLKYGAKPRPEKLPTPPPE 202
LLL ++ K PT P +
Sbjct 234 QSLLLSKISQDADLKTPTKPKQ 255
> CE19676
Length=597
Score = 32.3 bits (72), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 27/98 (27%), Positives = 43/98 (43%), Gaps = 1/98 (1%)
Query 94 NASKYTLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLE 153
N+ + + +A+ K ++E L+ L ALM A + E +Q ++E
Sbjct 280 NSEGFAPIHVAVRRLKLSLIEMLIEAGALIDFLDTEKKRNALMHAIEMNDFE-TIQLLVE 338
Query 154 RGANKSINDEDNEGRTALDRCEKGSPLYLLLLKYGAKP 191
RG+ +I DE E +L P+ LLL GA P
Sbjct 339 RGSGTNIEDESGETALSLAVKNVNYPVIGLLLDNGADP 376
> Hs16975496
Length=1663
Score = 32.3 bits (72), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 37/77 (48%), Gaps = 3/77 (3%)
Query 95 ASKYTLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLER 154
A + TLLQ A ++ LL ++ LD+N DG +AL +A + D V+ +L
Sbjct 708 AGRPTLLQQAAAQGNVTLLSMLLNEEGLDINYSCEDGHSALYSAA-KNGHTDCVRLLL-- 764
Query 155 GANKSINDEDNEGRTAL 171
A +N D G T L
Sbjct 765 SAEAQVNAADKNGFTPL 781
> Hs18252778
Length=587
Score = 32.0 bits (71), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 30/49 (61%), Gaps = 1/49 (2%)
Query 116 LLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLERGANKSINDED 164
L + D+NT+A+D +AL AC + E+ V+ +L +GA+ + ++D
Sbjct 241 FLAKYGADINTQASDNASALYEACKNE-HEEVVEFLLSQGADANKTNKD 288
> Hs4557799
Length=2321
Score = 32.0 bits (71), Expect = 1.3, Method: Composition-based stats.
Identities = 26/87 (29%), Positives = 44/87 (50%), Gaps = 6/87 (6%)
Query 88 DMNDDSNASKY---TLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVP 144
D D+NA + T L A+ + V + L+ + DL+ + DG+TAL+ A V
Sbjct 1860 DAGADTNAQDHSGRTPLHTAVTADAQGVFQILIRNRSTDLDARMADGSTALILAARLAV- 1918
Query 145 EDWVQAMLERGANKSINDEDNEGRTAL 171
E V+ ++ A+ +N D G++AL
Sbjct 1919 EGMVEELIASHAD--VNAVDELGKSAL 1943
> Hs14742395
Length=1194
Score = 32.0 bits (71), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 26/84 (30%), Positives = 44/84 (52%), Gaps = 3/84 (3%)
Query 88 DMNDDSNASKYTLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDW 147
D+N ++ + T L +A+ + +V+ LL D+N + +G+TALM A + E
Sbjct 1067 DVNAKASQAGQTALMLAVSHGRIDMVKGLLAC-GADVNIQDDEGSTALMCASEHGHVEIV 1125
Query 148 VQAMLERGANKSINDEDNEGRTAL 171
+ + G N + EDN+G TAL
Sbjct 1126 KLLLAQPGCNGHL--EDNDGSTAL 1147
> Hs10947052
Length=3957
Score = 32.0 bits (71), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 39/66 (59%), Gaps = 2/66 (3%)
Query 99 TLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLERGANK 158
T L +A + + +VV+ L+ ++ ++N ++ +G T L A Q+ D V+ +LE GAN+
Sbjct 99 TALHIASLAGQAEVVKVLV-KEGANINAQSQNGFTPLYMAA-QENHIDVVKYLLENGANQ 156
Query 159 SINDED 164
S ED
Sbjct 157 STATED 162
> Hs21264330_2
Length=500
Score = 31.6 bits (70), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 34/129 (26%), Positives = 57/129 (44%), Gaps = 10/129 (7%)
Query 89 MNDDSNASKYTLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWV 148
+ D +T L +A + ++V + L+ + D+N + + L A QQ V
Sbjct 171 LVDAKKEDGFTALHLAALNNHREVAQILIREGRCDVNVRNRKLQSPLHLAV-QQAHVGLV 229
Query 149 QAMLERGANKSINDEDNEGRTALD-RCEKGSPLYLLLLKYGAKPRPEKLPTP------PP 201
+++ G S+N ED EG TAL ++ L L+ G P P +L + P
Sbjct 230 PLLVDAGC--SVNAEDEEGDTALHVALQRHQLLPLVADGAGGDPGPLQLLSRLQASGLPG 287
Query 202 EPEVTVAAS 210
E+TV A+
Sbjct 288 SAELTVGAA 296
> 7301359
Length=1181
Score = 31.6 bits (70), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 26/82 (31%), Positives = 43/82 (52%), Gaps = 4/82 (4%)
Query 108 QKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLERGANKSINDEDNEG 167
++K+++E L + L LN K T L A + + D ++ +L++GA +N D+ G
Sbjct 377 KRKQLMELLTRKGSL-LNEKNKAFLTPLHLAA-ELLHYDAMEVLLKQGAK--VNALDSLG 432
Query 168 RTALDRCEKGSPLYLLLLKYGA 189
+T L RC + LLL Y A
Sbjct 433 QTPLHRCARDEQAVRLLLSYAA 454
> Hs10947054
Length=1872
Score = 31.6 bits (70), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 41/74 (55%), Gaps = 2/74 (2%)
Query 91 DDSNASKYTLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQA 150
D + T L +A + + +VV+ L+ ++ ++N ++ +G T L A Q+ D V+
Sbjct 91 DSATKKGNTALHIASLAGQAEVVKVLV-KEGANINAQSQNGFTPLYMAA-QENHIDVVKY 148
Query 151 MLERGANKSINDED 164
+LE GAN+S ED
Sbjct 149 LLENGANQSTATED 162
> At3g01750
Length=664
Score = 31.6 bits (70), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query 124 LNTKATDGTTALMTACDQQVPEDWVQAMLERGANKSINDEDNEGRTALD 172
+N + +G TAL A VP ++V+ ML + IN DN G T LD
Sbjct 335 VNYRNNEGRTALHLAISGNVPLEFVE-MLMSVKSIDINIRDNAGMTPLD 382
> Hs18601113
Length=108
Score = 31.2 bits (69), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 34/63 (53%), Gaps = 3/63 (4%)
Query 109 KKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPEDWVQAMLERGANKSINDEDNEGR 168
+ VV+ LL + D+N + DG+TALM AC+ E + +L + I+ D +G
Sbjct 8 RVDVVKALLACE-ADVNVQDDDGSTALMCACEHGHKE--IAGLLLAVPSCDISLTDRDGS 64
Query 169 TAL 171
TAL
Sbjct 65 TAL 67
> CE17508
Length=2761
Score = 31.2 bits (69), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 18/40 (45%), Positives = 25/40 (62%), Gaps = 6/40 (15%)
Query 139 CDQQVPEDWVQAMLERGANKSINDEDNEGRTALDRCEKGS 178
C QV V+ +L+RGAN + DED G+TALD+ + S
Sbjct 455 CLSQV----VKLLLQRGANPDLRDED--GKTALDKARERS 488
> Hs22051302
Length=389
Score = 31.2 bits (69), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 47/87 (54%), Gaps = 5/87 (5%)
Query 86 VSDMNDDSNASKYTLLQMALMLQKKKVVEYLLPQQDLDLNTKATDGTTALMTACDQQVPE 145
+ D+N ++ + T L +A+ ++ +V LL D+N + DG TALM A +
Sbjct 253 MGDVNAKASQTGQTALMLAISHGRQDMVATLL-ACGADVNAQDADGATALMCASEYGR-L 310
Query 146 DWVQAMLER-GANKSINDEDNEGRTAL 171
D V+ +L + G + +I DNEG +AL
Sbjct 311 DTVRLLLTQPGCDPAI--LDNEGTSAL 335
> At4g32500
Length=880
Score = 31.2 bits (69), Expect = 2.3, Method: Composition-based stats.
Identities = 20/46 (43%), Positives = 28/46 (60%), Gaps = 4/46 (8%)
Query 148 VQAMLERGANKSINDEDNEGRTALD-RCEKGSPL-YLLLLKYGAKP 191
+ +L+RG+N N+ D GRTAL KGS +LLL++GA P
Sbjct 558 LHQLLKRGSNP--NETDKNGRTALHIAASKGSQYCVVLLLEHGADP 601
Lambda K H
0.311 0.125 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5613892628
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40